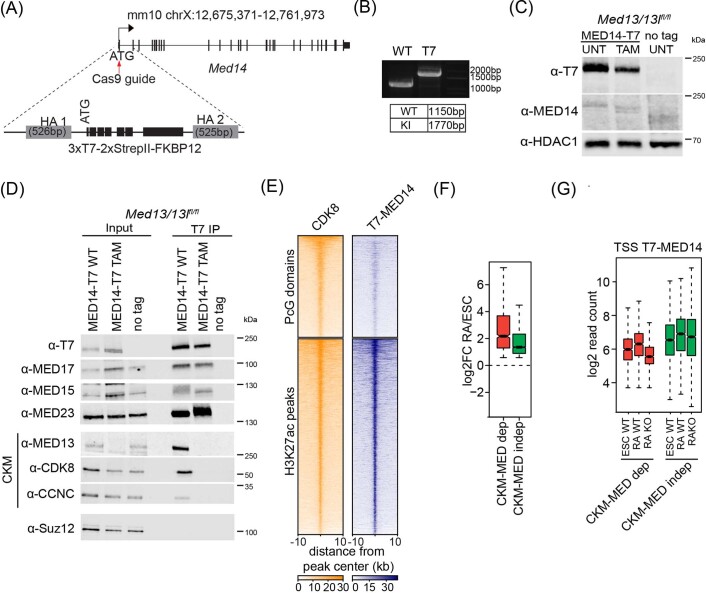
Extended Data Fig. 5. CKM-Mediator enables gene induction via recruitment of the Mediator complex.
a, A schematic illustration of the generation of the T7-MED14 expressing Med13/13lfl/fl ESC line. b, PCR showing amplification of homozygously-tagged T7-Med14 alleles (n = 2). c, A representative Western blot analysis of nuclear extracts from the T7-MED14 Med13/13lfl/fl ESC line, following tamoxifen (TAM) treatment (n = 3). Extract from an untagged ESC line was used as a control. HDAC1 was used as a loading control. d, A representative immunoprecipitation (IP) of endogenously T7-tagged MED14 with T7 antibody using nuclear extracts from Med13/13lfl/f ESCs before (UNT) and after tamoxifen (TAM) treatment (n = 2). The IPs were probed with the indicated antibodies. An IP from an untagged ESC line was performed as a negative control and a Western blot for SUZ12 was included as a control protein that does not interact with Mediator. e, Heatmaps of CDK8 and T7-MED14 ChIPseq signal at Polycomb domains (n = 2097) and H3K27ac peaks (n = 4037), sorted by decreasing CDK8 signal. f, Boxplots showing gene expression change (log2FC) of CKM-Mediator-dependent (n = 631) and CKM-Mediator-independent (n = 2689) RA-induced genes following RA differentiation of WT ESCs. Boxes show interquartile range, center line represents median, whiskers extend by 1.5x IQR or the most extreme point (whichever is closer to the median), while notches extend by 1.58x IQR/sqrt(n), giving a roughly 95% confidence interval for comparing medians. g, Boxplots showing T7-MED14 ChIPseq signal at the TSS (1000 bp) of the different classes of RA-induced gene classes as defined in e in ESCs and RA-induced cells (WT and CKM-Mediator KO). Boxes are defined as in f. Signal is an average from three independent biological experiments.