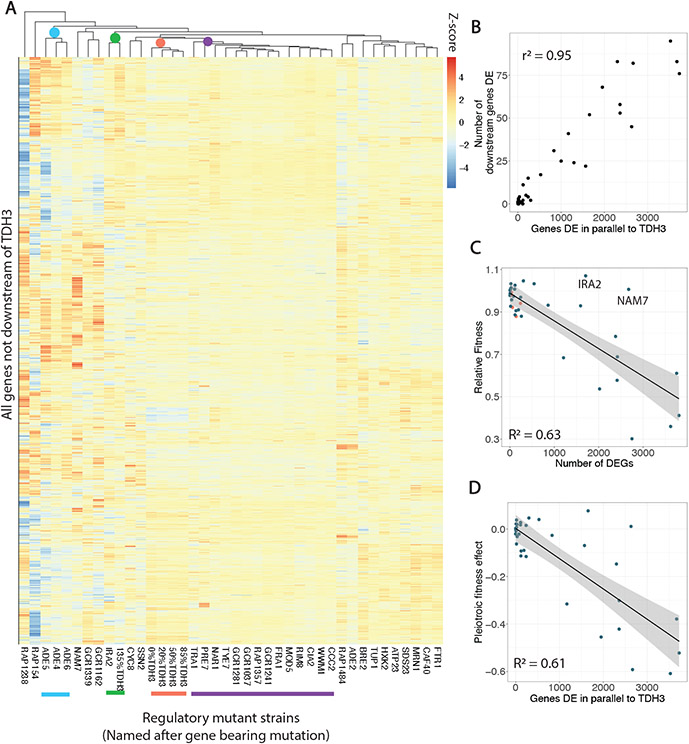
Fig. 4. Larger pleiotropic expression effects of trans-regulatory mutants correlate with negative fitness effects.
(A) A heatmap shows log2 fold changes for all genes not downstream of TDH3 as estimated by DESeq2, in which rows are genes and columns are mutants. Color intensity is scaled by row (by gene) and corresponds to z-scores. Hierarchical clustering of mutants is shown by the dendrogram above. Three of the four trans-regulatory mutants bearing mutations in the adenine biosynthesis pathway cluster together and are marked by a blue dot and line. The cis-regulatory mutant overexpressing TDH3 clusters with IRA2 (green), while the cis-regulatory mutants with reduced TDH3 expression cluster together (orange). A large cluster of trans-regulatory mutants with small effects across the genome are marked by a purple dot and line. (B) For all trans-regulatory mutants, the number of downstream genes differentially expressed is positively correlated with the number of genes differentially expressed in parallel to TDH3 in that mutant. (C) For both cis- (orange) and trans- (blue) regulatory mutants, the total number of differentially expressed genes is a strong predictor of relative fitness. A linear model is shown with grey shading representing 95% confidence intervals and two outliers, IRA2 and NAM7 are indicated. (D) For all trans-regulatory mutants with fitness data (i.e., excluding the two flocculant strains), the pleiotropic fitness effect, as defined in Fig. 2B, is plotted vs the number of genes differentially expressed in parallel to TDH3 in that trans-regulatory mutant. A linear regression is shown in black with 95% confidence intervals in grey.