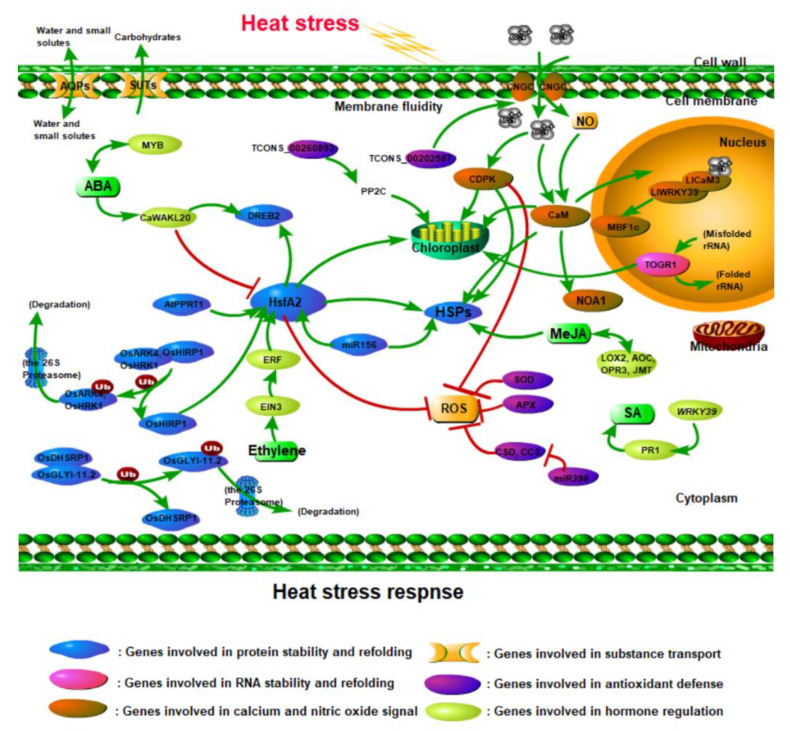
Figure 1.
Gene networks in plant heat stress response. The large regulatory networks involve genes related to protein and RNA stability and refolding, Ca2+, NO, and hormone signaling, substance transport, and antioxidant defense. Heat stress alters membrane fluidity which may activate Ca2+ channels resulting in an influx of Ca2+. Ca2+ signals are transduced by CaM (calmodulin) and CDPK (calcium-dependent protein kinase), activating signal transduction pathways in plants in response to heat stress. NO regulates the accumulation of HSPs (heat shock proteins) through AtCaM3. LlWRKY39 interacts with LlCaM3 in a Ca2+-dependent manner through the CaM-binding domain, and it promotes the expression of LlMBF1c. Under heat stress, HsfA2s are activated by AtPPRT1, OsHIRP1, ERF, and miR156. The expression of HsfA2 is activated to upregulate or fine-tune the expression of DREB2 (dehydration response element binding protein 2) and HSPs. As the main functional proteins induced by heat stress, HSPs and ROS (reactive oxygen species) constitute a complex regulatory network with CDPK, HsfA2, and some antioxidant enzymes. Both OsHIRP1 and OsDHSRP1 are degraded by the Ub/26S proteasome system to participate in the HSR (heat stress response) process. Under heat stress, TOGR1 expression is enhanced in the nucleolus, which helps the rRNA precursor to effectively fold. TOGR1, CDPK, and TCONS_00260893 protect chlorophyll synthesis under heat stress. AQPs (aquaporins) and SUTs (sucrose transporters) play important roles in maintaining the normal transport of water and sucrose under heat stress. In addition, some hormone-related genes are upregulated to promote hormone synthesis, such as MYB, EIN3, LOX2, AOC, OPR3, JMT, and WRKY39. Arrows denote the positive while red bars stand for negative interaction.