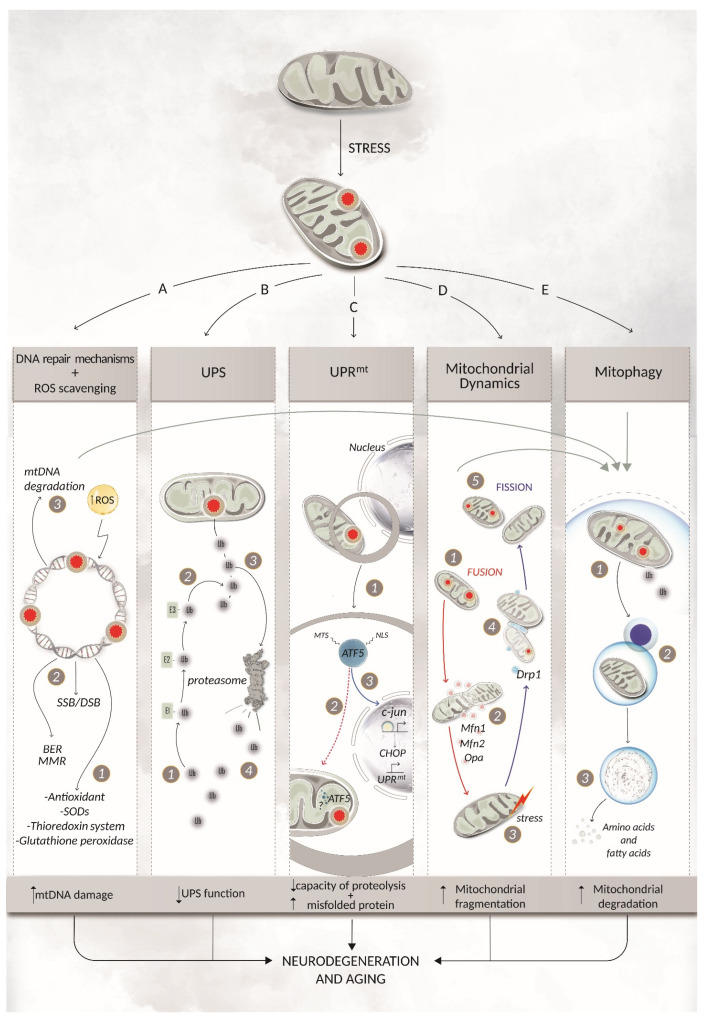
Figure 1.
Mitochondrial quality control: regulating mitochondrial turnover and homeostatic mtDNA repair mechanisms and ROS scavenging (A). Various mechanisms can be activated to either prevent or eliminate mtDNA damage. The first line of defence involves ROS scavenging factors such as antioxidants, SOD, the thioredoxin system, and glutathione peroxidase (1). Repair mechanisms become activated after damage is sensed on the mtDNA. Among these, the mitochondrial BER pathway is the most well-characterised, but in the last decades, other common proteins of nDNA repair mechanisms have also been identified in mitochondria, shedding light on the possibility that these pathways have a role in mtDNA repair as well (2) [13,14]. If all these damage responses are not able to fully repair the lesion, then a single molecule of mtDNA can be degraded. This does not impact organelle physiology since each mitochondrion owns multiple copies of the same nucleic acid (3). Finally, if the damage is extensive, the whole mitochondrion can be degraded through mitophagy. Ubiquitin proteasome system (UPS) (B). Dysfunctional mitochondrial proteins can be degraded by the UPS, a specific degradation system which relies on the covalent binding of ubiquitin to lysine residues within target proteins. Ubiquitin is translocated via the E1, E2, and E3 enzymes (1) before reaching the damaged protein (2). The polyubiquitin-tagged protein is then translocated to the cytosolic proteasome for degradation (3), where the ubiquitin is recycled, ready for another round of polyubiquitination (4). UPS is crucial in preserving mitochondrial integrity and vice versa. Indeed, dysfunctional mitochondria with an increased number of damaged proteins could not only overflow the proteasome but also affect the proteasomal subunits themselves, thereby affecting the catalytic activity of the UPS. Once mitochondrial dysfunction and proteasomal impairments develop, a vicious cycle may start, leading to a progressive failure of the UPS and, consequently, to ageing or, in the worst scenario, to neurodegenerative diseases. Mitochondrial unfolded protein response (UPRmt) (C). The UPRmt system can be activated in response to an accumulation of unfolded proteins in mitochondria. The crucial role of the UPRmt protein ATF5 is explicated through its nuclear localisation sequence (NLS) and mitochondria targeting sequence (MTS) (1). Under physiological conditions, ATF5 is localised in mitochondria (red dashed arrow) and likely degraded by a protease (2) such as the one characterised in C. elegans [15]. If mitochondrial import is dysfunctional, ATF5 accumulates in the cytosol and is translocated into the nucleus (blue arrow), where it can influence the activation of the transcriptional factors c-Jun and CHOP, which in turn regulate the activation of genes able to restore mitochondrial functions (3). Mitochondrial dynamics (D). Mitochondria can orchestrate cycles of fusion and fission as part of their dynamic network, allowing the maintenance of shape, distribution, and size. This mechanism can also be used to cope with unrepairable damages such as inter-/intrastrand and DNA-protein cross-links through the removal of the damaged section of the mtDNA by mitophagy [16]. Fusion ((1), red arrows) is mediated by Mfn1, Mfn2, and Opa1 (2) and allows the mitochondria to bond together to respond to damage (3). Fission (blue arrows) can also mediate the response to an external stress (3) that causes mitochondrial dysfunction. Fission is mediated by dynamin-related protein 1 (Drp1) (4) and, opposite to fusion, acts to isolate the damaged area of the organelle for clearance by mitophagy (5). Mitophagy (E). Defective mitochondria can be cleared in a process called mitophagy. The whole organelle is isolated from the rest of the cell owing to the generation of an autophagosome (1). The fusion of the autophagosome with a lysosome gives rise to the autolysosome (2) containing a set of enzymes in an acidic environment which drives the degradation of proteins, lipids, and nucleic acids in a controlled manner (3). BER = base excision repair; DSB = double strand break; MMR = mismatch repair; ROS = reactive oxygen species; SOD = super oxidase dismutase; Ub = ubiquitin; UPRmt = mitochondrial unfolded protein response; UPS = ubiquitin proteasome system.