Abstract
The yeast Saccharomyces cerevisiae has been used for bread making and beer brewing for thousands of years. In addition, its ease of manipulation, well-annotated genome, expansive molecular toolbox, and its strong conservation of basic eukaryotic biology also make it a prime model for eukaryotic cell biology and genetics. In this review, we discuss the characteristics that made yeast such an extensively used model organism and specifically focus on the DNA damage response pathway as a prime example of how research in S. cerevisiae helped elucidate a highly conserved biological process. In addition, we also highlight differences in the DNA damage response of S. cerevisiae and humans and discuss the challenges of using S. cerevisiae as a model system.
Keywords: DNA damage response, model system, Saccharomyces cerevisiae, yeast
1. Yeast: A Prime Eukaryotic Model System
Yeast has been indispensable to make beer and bread since 13,000–14,000 BC [1,2]. However, it was only in the 19th century that Louis Pasteur demonstrated the essential role of yeast in the fermentation process, where it is responsible for the conversion of cereal-derived sugars into ethanol and CO2 [3,4]. In 1883, Emil Christian Hansen was the first to isolate Saccharomyces pastorianus, the yeast responsible for fermentation of lager beers at the Carlsberg brewery [5]. Soon after, Saccharomyces cerevisiae was also isolated and quickly established itself as the most-commonly used yeast for the production of ale beers as well as a prime model for genetics and cell biology.
S. cerevisiae is a small (~5 µm) single-cell eukaryote and thus contains a nucleus and other membrane-bound organelles. Yeast cells are relatively easy to culture in laboratory conditions since they do not need a complex medium for growth. Moreover, cells divide rapidly, every 90 min, under optimal laboratory conditions through the process of budding, where a smaller, genetically identical daughter cell buds off the mother cell during mitosis. When, in 1978, Hinnen et al. published the successful transformation of yeast with a plasmid replicated in the bacterium Escherichia coli, yeast quickly became the most widely used single-cell eukaryotic model organism [6]. A plethora of replicating plasmids and selectable markers was developed. Moreover, apart from using plasmids, it quickly became clear that the efficient homologous recombination (HR) DNA repair machinery of S. cerevisiae allows integrating DNA fragments into specific genomic loci with an efficiency that is much higher compared to other organisms. Combined with the development of polymerase chain reaction (PCR), the efficient HR process enabled researchers to easily manipulate the yeast genome, which further established yeast as a model organism.
In 1996, the laboratory S. cerevisiae strain S288c became the first eukaryote for which the complete genome was sequenced. The results showed that a haploid S288c cell contains ~12,000 kilobases of genomic DNA, carrying approximately 6000 genes divided over 16 chromosomes [7]. This genetic architecture is specific for the S288c haploid lab strain that was sequenced. However, S. cerevisiae can exist in a haploid and diploid form. Moreover, naturally occurring S. cerevisiae strains are often genetically different and more complex, with a high degree of heterozygosity and frequently showing aneuploidy and polyploidy in their genomes [8,9,10].
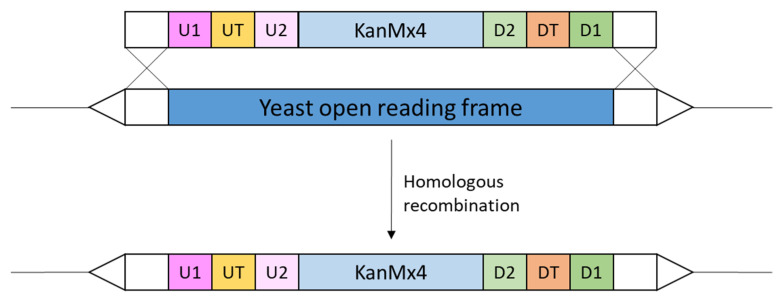
Soon after publication of the yeast genome sequence, the yeast deletion collection was constructed. This collection consists of a nearly complete set of viable deletion mutants where, in each deletion mutant, a specific non-essential open reading frame is replaced with a drug resistance marker flanked by two distinct 20 basepair sequences or DNA barcodes, the UPTAG and the DOWNTAG, which are unique for each open reading frame [11,12] (Figure 1). Each UPTAG and DOWNTAG is flanked by universal primer sequences to allow PCR amplification of the barcode. The high efficiency of homologous recombination in yeast was exploited to integrate the marker cassette and made the construction of this deletion collection possible. The yeast deletion collection was constructed in the haploid S288c strain and consists of approximately 4800 deletion mutants. Essential genes cannot be deleted in this haploid strain as this would be lethal therefore a heterozygous diploid yeast deletion collection was also constructed [12]. Moreover, deletion collections were also constructed in other S. cerevisiae strains such as the Sigma1278b strain [13]. Two yeast strains can differ as much as two human individuals, as such genes essential in one genetic background can be dispensable in another [13].
Figure 1.
Schematic overview of the construction of the yeast deletion collection. The cassette used to replace and thus delete every yeast open reading frame consists of a kanamycin-resistance gene (KanMx4) flanked by two 20 basepair sequences, called DNA barcodes, the UPTAG (UT) and the DOWNTAG (DT), which are unique for each gene. Each UPTAG and DOWNTAG is flanked by universal primer sequences (U1 and U2 for UPTAG, D1 and D2 for DOWNTAG) to allow PCR amplification of the barcodes. The DNA 5′ and 3′ to the barcodes is homologous to the yeast DNA flanking the yeast open reading frame (indicate by crosses). Yeast incorporates the cassette through homologous recombination which results in the replacement and thus deletion of the open reading frame by the cassette sequence.
The integration of DNA barcodes enables identification of individual mutants by their respective barcode, including tracking individual mutants in populations that contain a mix of all 4800 deletion mutants by deep-sequencing the barcode region from bulk samples, and then counting the relative proportion of each barcode [14,15,16,17,18]. The yeast deletion collection was crucial in research assigning specific cellular functions to genes. By growing the yeast deletion collection under different conditions, genes important for growth in particular conditions could be identified which significantly helped to assign the exact function of specific genes.
Several of the experimental procedures that were first developed in yeast cells, including barcoded siRNA screens [19,20] or CRISPR screens [21,22] were later also established in mammalian cells. However, these screens are often more complex compared to those in yeast. Other libraries such as the GFP library, in which each open reading frame is fluorescently tagged with GFP and a genome-wide over-expression library were constructed in S. cerevisiae to further characterize gene functions [23,24]. Furthermore, transcriptome analysis using DNA microarrays was also developed in yeast [25,26]. This was followed by more omics technologies such as proteomics and metabolomics as well as the development of methods such as ChIP-seq to map transcription factor binding [27,28,29,30,31].
Together, the molecular toolbox that was developed in S. cerevisiae and expanded to other organisms much accelerated research in cell biology, genetics, and genomics. Importantly, all information is collected and organized in the Saccharomyces Genome Database (SGD), a freely available online database which provides information on DNA and protein sequence, expression, regulation, interactions, phenotypes, Gene Ontology (GO) annotations, etc. for every yeast gene [32,33].
In 1987, it was shown that a human gene could complement a yeast mutant defective in the cell cycle [34]. This was a major breakthrough, since it illustrated that cell cycle control mechanisms were highly conserved between yeast and humans. In fact, despite being separated by approximately 1 billion years of evolution, more than one-third of the yeast genome has a homologous part in humans [35]. Furthermore, comparing the yeast genome sequence to that of other model organisms such as Caenorhabditis elegans, Drosophila melanogaster and the human genome sequence showed that protein amino acid sequences and protein functions have been conserved to such extent that GO annotations could frequently be transferred from one eukaryotic species to another. On average 32% amino acid identity is detected between yeast and human over the complete genome [36]. Additionally, approximately 50% of genes essential in yeast can be replaced by their human homolog [36].
The characteristics of yeast discussed above, namely the short life cycle, ease of manipulation, well-annotated genome, expansive molecular toolbox along with the strong conservation of basic eukaryotic biological and biochemical pathways, make yeast an excellent model organism to study eukaryotic cellular processes. Consequently, the yeast model has been used extensively in many fields, some of which will be shortly illustrated here.
There are many examples of key discoveries in the field of biology, genetics, biochemistry and medicine that have been made in yeast cells. A prime example is the discovery of key regulators of the cell cycle, for which the Nobel Prize was awarded in 2001 to Leland H. Hartwell, Tim Hunt and Sir Paul M. Nurse. Hartwell used S. cerevisiae to identify more than one hundred genes involved in cell cycle control, the cell division cycle or CDC genes [37,38,39]. He found that one of these genes, CDC28, controlled the first step in the progression through the G1 phase. He also introduced the concept of checkpoints as he discovered that the cell cycle is arrested upon DNA damage induced by X-ray radiation to allow time for repair [40]. This was later extended to multiple checkpoints to ensure a correct order of cell cycle phases [41,42,43,44]. Sir Paul M. Nurse used the yeast Schizosaccharomyces pombe as a model to identify the gene CDC2 which was identical to CDC28 identified by Hartwell [45,46,47,48,49]. The basic discoveries in yeast cells led to the hypothesis that defects in cell cycle checkpoints could be responsible for the uncontrolled growth and genomic instability of cancer cells [50]. An increased understanding of the cell cycle shed light on the molecular mechanisms for cellular transformation from normal to cancer cells which helped to identify targets for cancer therapies [51,52].
Other key eukaryotic cellular processes such as eukaryotic transcription, telomere structure, vesicle transport and autophagy were also unraveled using yeast as a model system, and several of the pioneering scientists were awarded Nobel prizes. In 2006, Roger Kornberg received the Nobel Prize in Chemistry “for his studies of the molecular basis of eukaryotic transcription” for deciphering the structure of the key components for transcription in yeast [53,54,55,56,57,58,59,60]. Jack Szostak, Elizabeth Blackburn, and Carol Greider used yeast to discover the telomerase function in eukaryotes and have received the Nobel Prize in Physiology and Medicine in 2009 [61,62,63,64,65,66,67,68,69,70]. In 2013 James E. Rothman, Randy W. Schekman and Thomas C. Südhof received the Nobel Prize “for their discoveries of machinery regulating vesicle traffic, a major transport system in our cells”. Schekman et al. used yeast to identify mutated genes that caused a defect in the transport machinery [71,72,73,74,75,76,77]. He then established an ordered secretory system and further elucidated the underlying mechanism. Later Rothman discovered a protein complex in mammalian cells that enables vesicles to dock and fuse with their target membranes [78,79]. Strikingly, some of the genes Schekman had discovered in yeast were homologous to those Rothman identified in mammals. This again demonstrated the conservation of a basic mechanism from yeast to man. In 2016 the Nobel Prize in Physiology or Medicine was awarded to Yoshinori Ohsumi “for his discoveries of mechanisms for autophagy” [80,81,82]. Ohsumi was the first to identify multiple yeast genes that regulate autophagy.
Yeast also serves as an important model for medical research. Almost 30% of known genes involved in human disease have yeast homologs [83,84]. Genome-wide approaches have been especially useful to screen for new biologically active compounds and to unravel drug-induced molecular mechanisms. The yeast deletion collection has been used to screen many different drugs to identify their molecular targets, including for example molecules that limit tumor growth such as Wortmannin and 5-Fluorouracil [85,86,87,88,89,90,91,92]. The heterozygous diploid yeast deletion collection has been useful in particular to screen for drug-induced haploinsufficiencies. This assay is based on the observation that a heterozygous deletion strain is sensitive to a drug that targets the protein expressed from the heterozygous locus [85]. The power of this assay lies in the simultaneous identification of both the inhibitory compound and its targets without prior knowledge of either of the two. The haploid yeast deletion collection cannot be used to identify targets of a certain drug, because the target is absent. However, the deletion collection is particularly useful for identifying genes that act to buffer the compound target pathway and are thus required for growth in presence of the compound. As a result, this assay can be used to screen compounds that do not directly target a protein, such as DNA damaging agents or reactive oxygen species (ROS)-inducing compounds, and identify genes important for the response to these agents [14,93].
Central to all these discoveries is the high conservation of basic biological processes between yeast and humans. One of such highly conserved pathways is the DNA damage response, which makes it possible to use yeast as model system to increase our basic understanding of the DNA damage response. In the following sections, the highly conserved DNA damage response of S. cerevisiae is described and compared to humans.
2. The DNA Damage Response in S. cerevisiae and Humans
DNA is susceptible to damage by endogenous as well as external factors such as ultraviolet (UV) light, ionizing radiation, and alkylating agents. Damage induced in the DNA can be in the form of single-strand breaks (SSB) and double-strand breaks (DSB). DSBs are considered most harmful since this type of lesion is less efficiently repaired. Upon DNA damage, the DNA damage response is activated to repair the DNA and promote cell survival. In the next sections, the DNA damage response and the major SSB and DSB DNA repair pathways of S. cerevisiae are described.
The core DNA repair process of the different pathways are largely similar, with 70% of yeast DNA repair proteins having a human homolog. However, DNA repair mechanisms in humans often involve a larger number of proteins and components. Here, we do not describe human DNA repair pathways in full detail, but homologs of the yeast proteins are shown in the figures and relevant differences in DNA repair between yeast and humans are highlighted. Examples illustrate how S. cerevisiae has been used as a model organism.
2.1. Signaling of DNA Double-Strand Breaks
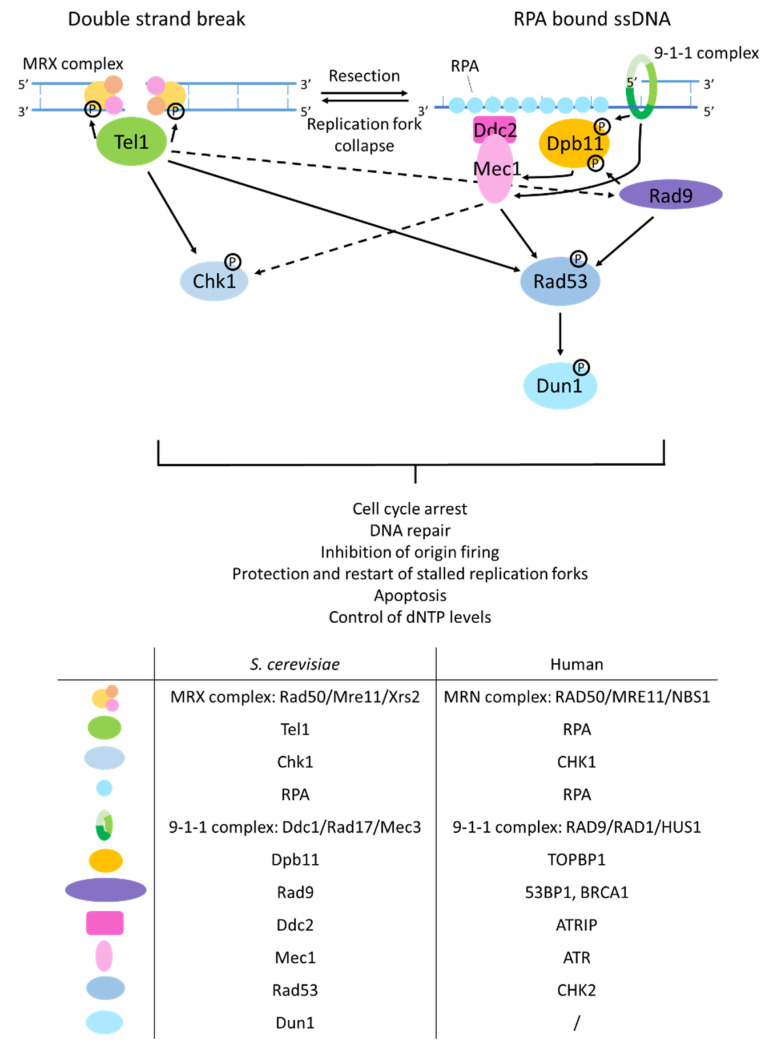
In S. cerevisiae, signaling of DNA damage depends on the kinases Mec1 and Tel1 (Figure 2). DSBs are recognized by the MRX complex (composed of Mre11, Rad50 and Xrs2), which recruits and activates Tel1 [94,95]. Tel1 in turn activates the effector kinase Chk1. Mec1 is recruited to stretches of ssDNA bound by replication protein A (RPA, a heterotrimeric protein formed by the subunits Rfa1, Rfa2 and Rfa3) through its interaction partner Ddc2, where it phosphorylates and activates Rad53 together with Dpb11 which is loaded onto ds/ssDNA junctions by the 9-1-1 complex, composed of the proteins Ddc1, Rad17, and Mec3. Both Tel1 and Mec1 can phosphorylate Chk1, Rad9, and Rad53 directly and can as well phosphorylate lesion-proximal substrates such as histone H2A. Rad9 can bind these histone modifications and serve as an adaptor protein for Rad53 activation [96,97]. The activated DNA damage signaling kinases Chk1 and Rad53 then mediate the responses to DNA damage that include cell cycle arrest, activation of DNA repair pathways, inhibition of origin firing, protection and restart of stalled replication forks, initiation of apoptosis and control of dNTP levels [98,99,100]. Rad53 can further phosphorylate and activate Dun1, which upregulates the transcription of a specific set of DNA damage-induced genes, including subunits of ribonucleotide reductase (RNR) [99,101,102]. Human homologs of the sensing kinases Mec1 and Tel1 are ATR and ATM, respectively. Their function and phosphorylation cascade are largely similar to the process in yeast; however, a DUN1 homolog has not been identified. Instead activation of ribonucleotide reductases is performed by p53, which is a target of the checkpoint kinases CHK1 and CHK2, the mammalian homologs of Chk1 and Rad53 [103].
Figure 2.
DNA damage signaling pathways. Upon DNA damage two sensor kinases Tel1 and Mec1 start a phosphorylation cascade. Double-strand breaks (DSB) are recognized by the MRX complex, which in turn is recognized by Tel1. Tel1 then phosphorylates Chk1. ssDNA stretches bound by RPA are recognized by Ddc2. Together the 9-1-1 complex and Dpb11 activate Ddc2-Mec1. Activated Mec1 phosphorylates Rad53, which in turn phosphorylates Dun1. Rad9 serves as an adaptor protein to activate Rad53. Crosstalk between Tel1 and Mec1 can occur when a stalled replication fork collapses which can result in a DSB or when a DSB is resected which results in ssDNA stretches. Human homologs of the different yeast proteins are listed in the table.
2.2. Single-Strand Break Repair Mechanisms
2.2.1. Base Excision Repair
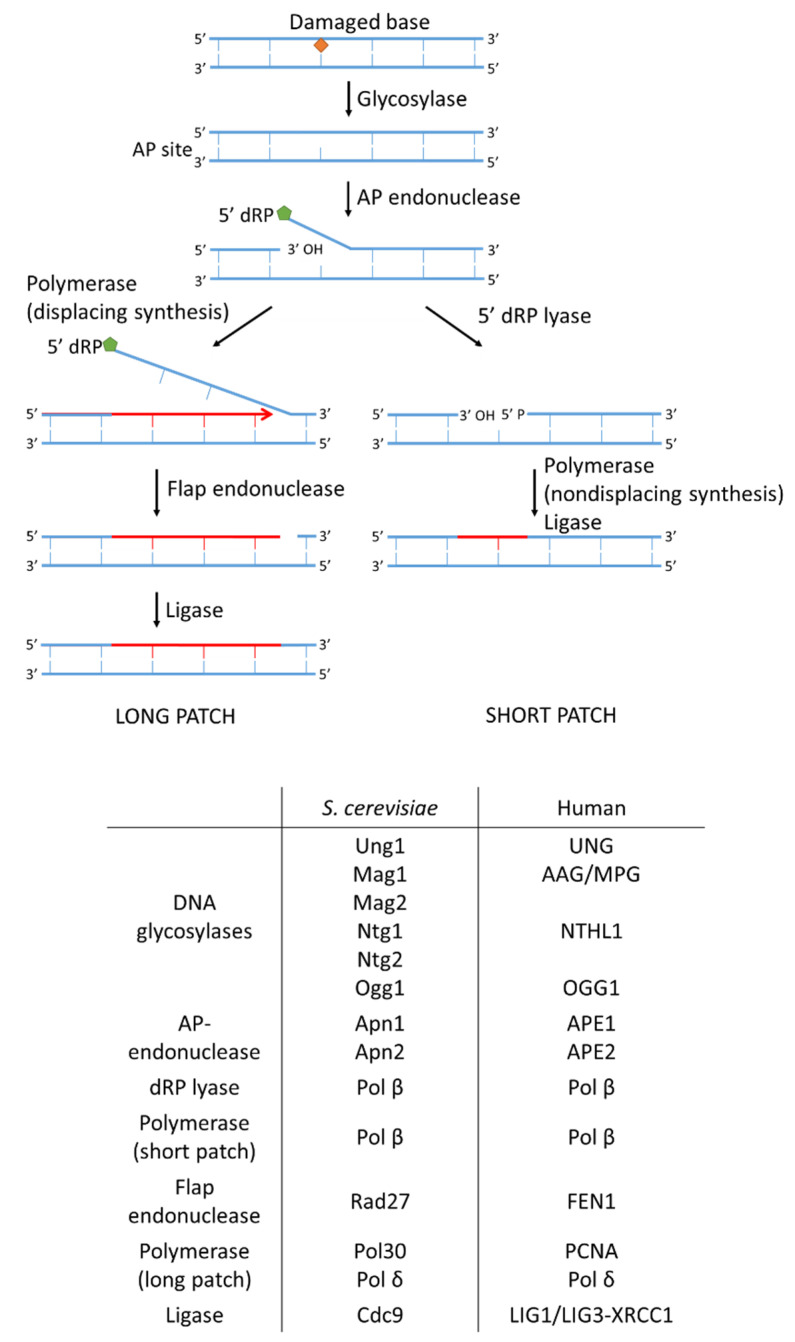
DNA lesions that are not associated with structural alterations of the DNA helix such as oxidized, deaminated and alkylated bases or apurinic/apyrimidinic sites (AP sites) are repaired by base excision repair (BER) [104]. In S. cerevisiae, BER occurs in two stages (Figure 3). First, a damage-specific step depends on DNA glycosylases that recognize a specific type of base lesion. S. cerevisiae contains six DNA glycosylases: monofunctional Ung1, Mag1, and Mag2 and bifunctional Ntg1, Ntg2, and Ogg1, each targeting specific damage [105,106,107,108,109]. For example, Mag1 recognizes alkylated adenine bases, whereas Ogg1 targets 8-oxoguanine. The glycosylases bind the minor groove of the DNA, thereby inducing a kink. Next they flip the abnormal base out of the DNA helix and cleave the β glycosidic bonds between the ribose and base [110]. The resulting 3′ deoxyribose phosphate is then processed by AP endonucleases [111]. In S. cerevisiae two AP endonucleases are known; Apn1 and Apn2 [112,113]. The 3′ OH site serves as the initiation site for DNA synthesis and the 5′ diphosphate (5′-PP) end facilitates DNA polymerase binding [114].
Figure 3.
Mechanisms of base excision repair (BER), mismatch repair (MMR), and nucleotide excision repair (NER). (A) Mechanism of BER. The base lesion is recognized and removed by DNA glycosylases. The resulting apuric or apyrimidic (AP) site is processed by AP endonucleases. In a long patch, BER displacing synthesis is followed by flap removal by Rad27, and ligation takes place. In a short patch, BER deoxyribose phosphate (dRP) lyase excises one nucleotide, which is followed by gap filling and ligation.
There are two subpathways of BER: short and long patch repair. In short patch repair, one single nucleotide is excised. Gap filling and ligation is performed by polymerase β and the ligase Cdc9. In long patch repair, polymerase δ and Pol30 are responsible for gap filling with strand displacement resulting in 2–13 nucleotide being replaced [115]. This process creates a 5′ overhang tail, which is cleaved by the flap endonuclease Rad27 [116]. The BER pathway is largely similar in humans; homologs of the yeast proteins are listed in Figure 3. In addition, an important additional protein, poly(ADP-ribose)polymerases 1 (PARP-1), functions in human BER but does not have a yeast homolog [117]. PARP-1 recognizes and binds to DNA SSBs and recruits other BER repair proteins such as DNA [118,119,120] polymerase β and the XRCC1-DNA ligase III complex to the site of damage [121].
As BER was first described in E. coli, yeast has proven particularly useful in the translation of the BER pathway from a prokaryotic to a eukaryotic system. This is illustrated here with example of the identification of OGG1. In E. coli, the genes Mutm and Fpg are two DNA glycosylases that prevent spontaneous mutagenesis [122]. Consequently, inactivation of both mutM and Fpg results in a mutator strain which was used to identify the OGG1 gene from S. cerevisiae in a functional complementation assay. Therefore, the E. coli strain lacking mutM and Fpg was transformed with a yeast DNA library and clones that showed a reduced spontaneous mutagenesis were selected. Subsequent sequencing and characterization identified the OGG1 gene on chromosome XIII [105]. Scanning the human sequence databases for homology to the S. cerevisiae OGG1, the human OGG1 gene was rapidly identified. A similar approach using a human genomic library would not have been possible, as the Ogg1 protein had no sequence similarity to the E. coli Fpg or MutM. As such cloning strategies based on sequence similarity would not have been successful and a functional complementation assay was necessary to identify OGG1. However, in E. coli functional complementation is less successful for human genes as E. coli lacks the mRNA splicing machinery. S. cerevisiae on the other hand harbors relatively few introns. As such a yeast genomic library can be used for expression in E. coli. This example illustrates the power of yeast as tool to identify the BER genes.
2.2.2. Mismatch Repair
Ensuring correct DNA replication is essential for maintaining genomic stability. It has been estimated that a wrong nucleotide is incorporated once every 104 to 105 nucleotides [123]. However, the observed final error rate after DNA replication is much lower, 0.33 × 10−9 per site per cell division for S. cerevisiae and 0.1 × 10−9 per site per cell division for humans [124]. This suggests the existence of replication and post-replication DNA repair mechanisms that correct part of the erroneous incorporations. Firstly, high-fidelity DNA polymerases possess proofreading activity that can remove wrongly incorporated bases. Second, DNA mismatch repair (MMR) is a highly conserved DNA repair pathway that corrects mismatches that have escaped the proofreading activity of the polymerases. The MMR repair pathway was extensively studied in Escherichia coli and homologs were later identified and further characterized in eukaryotes, including yeast [125,126].
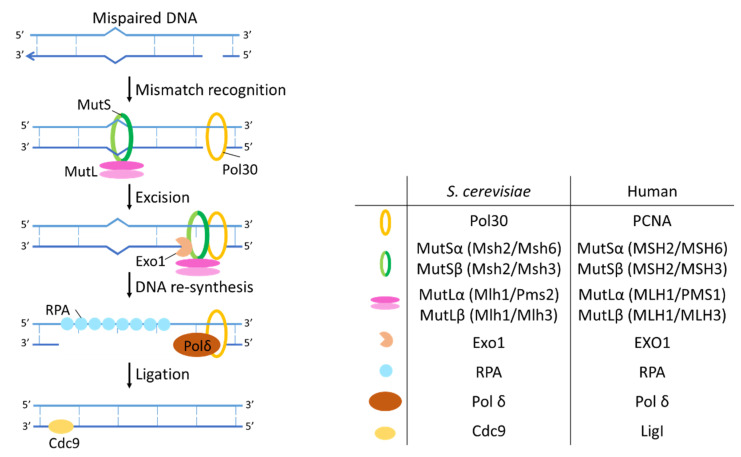
In S. cerevisiae, the different steps in the mismatch repair pathway include mismatch recognition by the yeast MutS and MutL homologs [127,128,129], excision of a region that contains the mismatched base by the exonuclease Exo1 [130,131,132] and DNA re-synthesis followed by ligation (Figure 4). In eukaryotes MutS is a heterodimeric protein that can consist of either Msh2/Msh6 (MutSα) or Msh2/Msh3 (MutSβ). Substrate recognition by the MutS homologs is followed by a conformational change of MutS homologs driven by ATP hydrolysis [133,134]. MutS changes into a sliding clamp and recruits the heterodimer of MutL homologous [135]. Here too there are several MutL homologs: MutLα, consisting of Mlh1/Pms2 and MutLβ formed by Mlh1/Mlh3. The complex that is now formed by MutS and MutL homologs can slide over the DNA in both directions due to the sliding clamp activity of MutS. MutL is essential for the recruitment of downstream MMR factors to the lesion [136].
Figure 4.
Mechanism of MMR. MutS and MutL homologs recognize the mismatch followed by excision of the mismatch by Exo1 and DNA re-synthesis and ligation.
The sliding continues until the complex encounters a strand discontinuity or nick (for example a gap between Okazaki fragments) that is bound by the DNA polymerase processivity factor proliferating cell nuclear antigen (PCNA) encoded by POL30 in yeast [137,138]. PCNA is important to link the MMR complex to DNA polymerase at the replication fork, ultimately facilitating the distinction between daughter and template strand [139]. When MutS-MutL encounters PCNA, this initiates loading of the exonuclease Exo1 [130,131,132]. Exo1 then starts degrading the nicked strand and thereby removes the mismatch. The single stranded gap in the DNA is stabilized by RPA and filled in by polymerase δ using the parental DNA strand as a template. Lastly the DNA is ligated by Cdc9 in [140,141].
2.2.3. Nucleotide Excision Repair
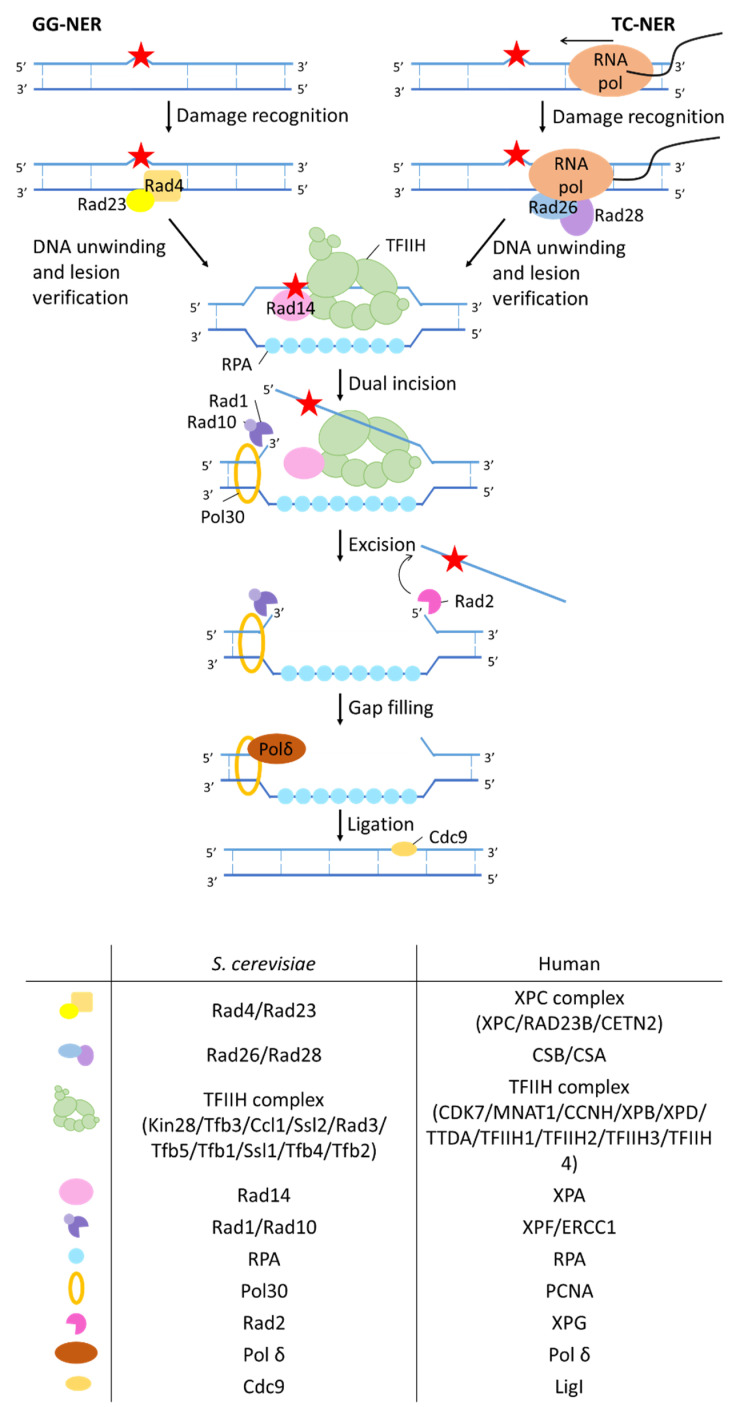
Nucleotide excision repair (NER) is a DNA repair mechanism that recognizes and repairs bulky DNA damage (Figure 5). Depending on where in the genome the lesion is located, two different subpathways of NER recognize the lesion, namely global genome nucleotide excision repair (GG-NER) and transcription-coupled nucleotide excision repair (TC-NER). In GG-NER damages in the entire genome, including non-transcribed regions and silent chromatin, are repaired. TC-NER on the other hand is responsible for the repair of lesions in the transcribed strand of active genes.
Figure 5.
Mechanism of NER. In global genome (GG) NER, the Rad4/Rad27 complex recognizes the lesion. In transcription-coupled (TC) NER, the Rad26/Rad28 complex recognizes the stalled RNA polymerase. Next the transcription initiation factor IIH (TFIIH) complex is recruited. The helicase Ssl2 and Rad3 within the TFIIH complex extend the open DNA configuration around the lesion with their opposite helicase polarity. Rad14 stimulates TFIIH helicases activity while RPA stabilizes the ssDNA by binding to it. Dual incision is performed by two structure-specific endonucleases Rad2 and Rad10-Rad1 which is followed by gap filling and ligation. Human homologs of the different yeast proteins are listed in the tables.
In GG-NER, the Rad23-Rad4 complex continuously scans the DNA until a lesion is recognized [142]. When the Rad23-Rad4 complex actually recognizes a damaged DNA site, a more stable Rad23-Rad4-DNA complex is formed and downstream NER factors are recruited to the site of damage.
As the name suggests, transcription coupled (TC) NER is initiated when damage to a transcribed DNA strand limits transcription activity of that strand. The stalled RNA polymerase serves as DNA damage recognition signal during transcription. The lesion-stalled RNA polymerase is recognized by the TC-NER-specific protein Rad26, which then recruits Rad28 that is required for further assembly of the NER complex [143,144,145].
After damage recognition, the transcription initiation factor IIH (TFIIH) complex together with the proteins RPA and Rad14 are recruited to the DNA damage site in both TC-NER and GG-NER. TFIIH is a multisubunit complex composed of two helicases (Ssl2 and Rad3), the complex of CDK-activating kinase (CAK: Kin28 and Ccl1) and structural proteins (Tfb1, Tfb2, Tfb3, Tfb4, Tfb5 and Tfb6) that form the core. The structural proteins have no enzymatic activity. The two helicases have opposite polarities (Ssl2 3′–5′, Rad3 5′–3′), which results in an extension of the open DNA configuration around the lesion, forming a DNA bubble. Rad14 stimulates TFIIH helicases activity, while RPA stabilizes the ssDNA by binding to it. On top of its helicase activity, Ssl2 also possesses ATPase activity, which enables the recruitment of the TFIIH factor [145,146,147].
Next, dual incision is performed by two structure-specific endonucleases Rad2 and Rad10-Rad1. The endonucleases respectively cut the damaged strand 3′ and 5′ from the lesion, leaving a single-strand gap of 22–30 nucleotides [145,148,149].
RPA and Rad2 further coordinate the synchronization of lesion excision and DNA gap filling, which prevents the accumulation of ssDNA gaps that may induce DNA damage signaling. Directly after the 5′ incision Pol30 (PCNA) is loaded and recruits a DNA polymerase (DNA Pol δ, DNA Pol κ or DNA Pol ε) to fill the gap in the DNA [150]. Finally, DNA ligase Cdc9 seals the nick [151].
In humans GG-NER is performed by the XPC complex (composed of XPC, Rad23b, and Cetn2). However, the XPC complex is inefficient in recognizing the typical lesions induced by UV light exposure, namely cyclobutane pyrimidine dimers, due to their low degree of structural perturbation [152]. Consequently, these lesions are recognized by an alternative complex, namely the UV-DDB complex. This complex is composed of two DNA damage binding proteins, DDB1 and DDB2. These two proteins associate with the CRL complex composed of CUL4A and RBX1 (ROC1). The newly formed complex aids in remodeling the damaged site to prepare it for subsequent NER reactions by ubiquitination of histones H3, H4 and H2A [153,154,155]. These ubiquitination events enhance the recruitment of XPC to the site of damage and thereby facilitate the repair process. DDB2 binds to the UV-induced damaged DNA, extrudes the lesion into its binding pocket and kinks the DNA [156]. This action of DDB2 creates ssDNA, which facilitates XPC binding.
NER has been extensively studied in mammals as deficiency in NER is associated with multiple syndromes such as xeroderma pigmentosum and Cockayne syndrome. As such studies in cell lines of xeroderma pigmentosum patients have enhanced our knowledge about NER [157]. Nonetheless, genetic and biochemical studies in yeast made major contributions in elucidating the molecular mechanisms of NER. For example, studies with a rad1 mutant allele, which encoded a protein that could not interact with Rad10, revealed that complex formation was essential for the functioning of these proteins [158].
2.3. Double-Strand Break Repair Mechanisms
2.3.1. Homologous Recombination
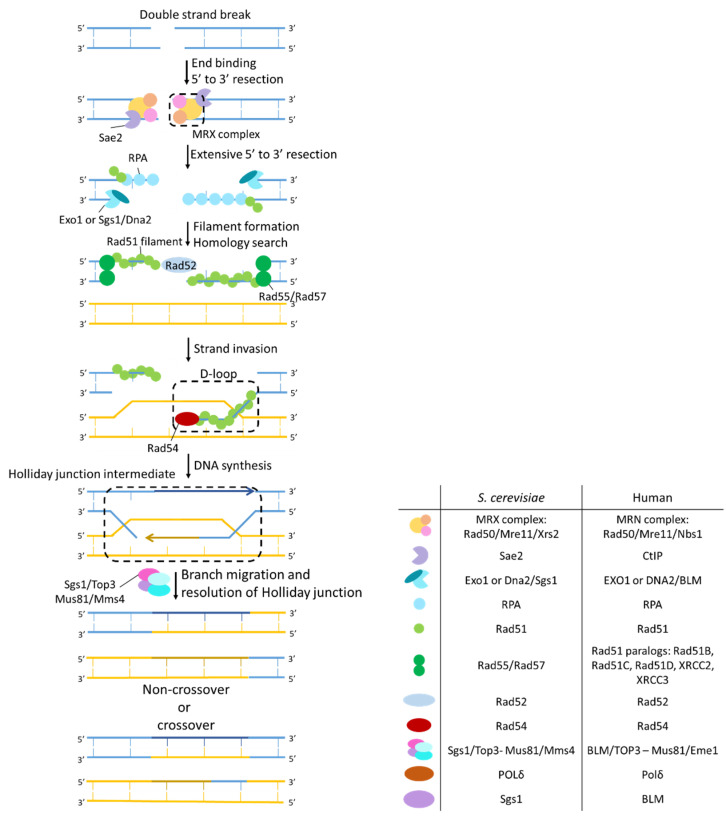
HR is a DSB repair pathway conserved from bacteria to humans. In yeast it the most-used DSB repair pathway (Figure 6). This DNA repair pathway allows accurate, high-fidelity repair of DSBs because it uses the undamaged sister chromatid as a repair template. As yeast transformations rely on HR, this pathway was extensively studied in this model organism, thereby using the unique properties of the MAT locus and HO endonuclease.
Figure 6.
Mechanism of homologous recombination (HR). DSBs are recognized by the MRX complex. Sae2 catalyzes initial end processing, after which Exo1, Sgs1, and Dna2 perform extensive end resection to form long stretches of ssDNA bound by RPA. Rad51 mediated by Rad52 and Rad55-Rad57 replaces RPA to form the Rad51 filament. Rad54 helps the Rad51 filament to find homologous sequences. When a homologous sequence is found, strand invasion is performed forming the displacement loop or D-loop. Engaging the second end of the DSB mediated by Rad52 forms a double Holliday junction. The double Holliday junction is resolved by the combined action of the helicase Sgs1, the topoisomerase Top3, or Mus81 and Mms4, resulting in crossover and non-crossover products.
Depending on the allele present in the MAT locus, haploid S. cerevisiae can exist in two mating types, a or α (diploid cells typically carry one a and α allele). Via homologous recombination initiated by a DSB at the MAT locus, haploid yeast cells can autonomously change mating type. The introduction of this DSB is catalyzed by the endogenously encoded HO endonuclease. Mating type switching then occurs via homologous recombination using the opposite mating type information, present at the HML and HMR loci, as a template [159]. In normal conditions, the HO gene is tightly regulated. However for DSB repair studies, a galactoses-inducible HO gene was engineered [160]. This enabled to express HO in all cells at the same time and thus to introduce a DSB at the MAT locus to all cells simultaneously by growing the yeast cells on galactose [161,162]. Further engineering the recognition sequences allowed to introduce a DSB at different loci in the genome, which facilitated the unraveling of the mechanistic details of the process [163,164,165]. Furthermore, by analyzing the repair kinetics and intermediates in strains where genes involved in homologous recombination were mutated or deleted, the role of these genes in homologous recombination could be investigated [166,167,168]. Moreover, immunostaining and chromatin immunoprecipitation (ChIP) protein recruitment to a double-strand break could be assayed [169,170,171,172].
The model for repairing broken ends using a homologous template was first proposed by Resnick in 1976 [173]. In a first step, the DSB is pre-processed to a 3′ overhanging tail by extensive DNA end resection [174]. First, initial end processing is performed by the MRX complex (Mre11, Rad50 and Xrs2) together with Sae2, followed by more extensive resection by the exonuclease Exo1 or the helicase Sgs1 together with the nuclease Dna2 [175,176]. The resulting single stranded DNA (ssDNA) is bound by RPA, a ssDNA stabilizing protein. Next, Rad51 needs to replace RPA in order to form the Rad51 filament. As RPA is an abundant protein and binds ssDNA with very high affinity, recombination mediator proteins, Rad52 and Rad55-Rad57, are needed to help Rad51 replace RPA [172,177,178,179,180]. When Rad51 is completely loaded onto the ssDNA, the presynaptic filament is formed.
The presynaptic filament, facilitated by Rad54, scans the genome for sequence complementarity in order to find a homologous part from which to initiate DNA synthesis [181,182]. When a homologous donor dsDNA is found, the synaptic complex is formed. The intermediate that is formed by strand invasion is called a displacement loop or D-loop, because the original base pairs in the donor dsDNA must be disrupted and replaced by base pairing of the invading strand with the donor. The D-loop can migrate in the DNA via replication by DNA polymerase δ [183]. Before DNA synthesis can start, Rad51 must be dissociated from the heteroduplex DNA. This is mediated by Rad54 [172,181,182]. Next, the second end of the DSB is engaged by Rad52 in a process called second end capture, which results in the formation of a double Holliday junction [184,185,186]. The double Holliday junction is resolved by the combined action of the helicase Sgs1, the topoisomerase Top3 or the Mus81/Mms4 complex resulting in crossover and non-crossover products [187,188,189]. In crossover products the part of the homologous strand used as a template is exchanged. In non-crossover products this is not the case.
In humans, the MRN complex together with CtIP initiate limited end resection which is further extended by BLM helicase together with EXO1 and DNA2 exonucleases. RPA than binds to the single stranded DNA ends and is replaced by Rad51, which in humans in mediated by BRCA2.
2.3.2. Non-Homologous End-Joining
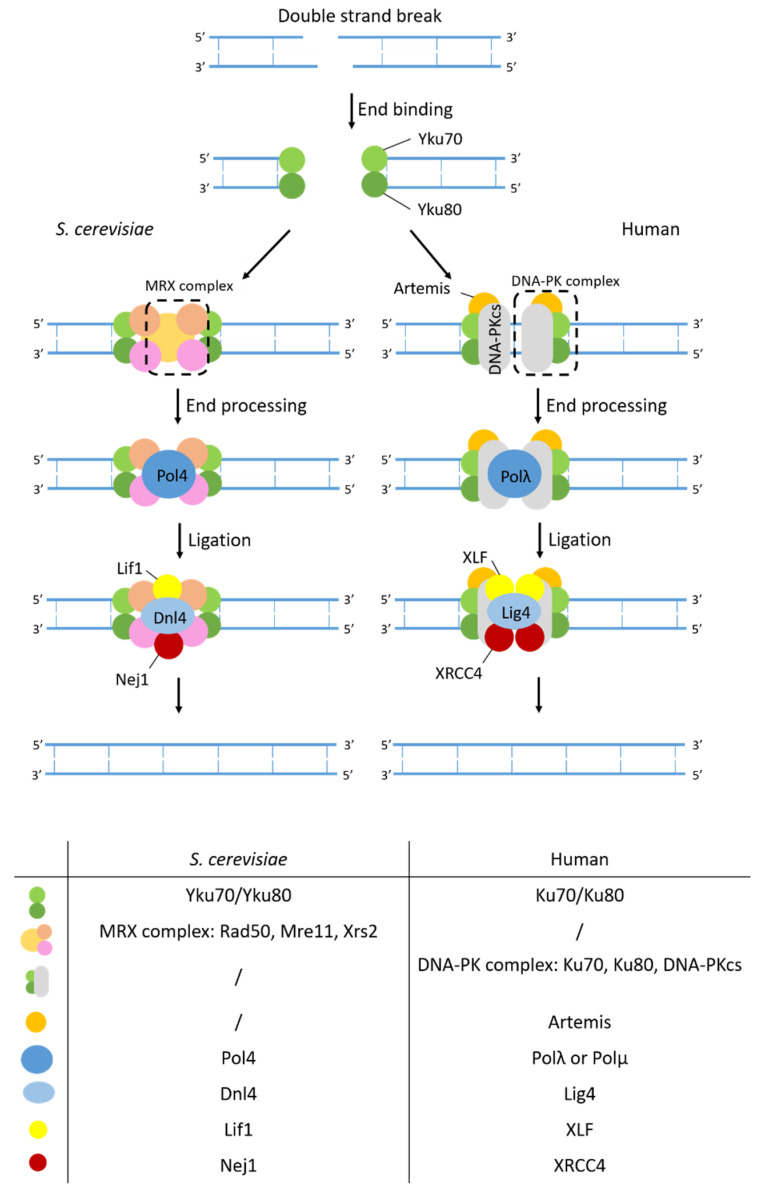
Non-homologous end-joining (NHEJ) is a second type of DSB repair pathway that directly ligates the DNA ends together in an error-prone manner (Figure 7). Contrary to yeast, where HR is the most used DSB repair pathway, in humans NHEJ is the most common pathway to repair DSBs.
Figure 7.
Mechanism of NHEJ. In yeast NHEJ, Yku70 and Yku80 bind the DSB ends together with the MRX complex. The Ku complex recruits other NHEJ proteins such as ligase Dnl4-Lif1 and the stabilizing factor Nej1 forming the repair super complex. Pol4 works together with the repair super complex making complementary ends which are ligated by Dnl4. In humans the process of NHEJ is largely similar, although the MRX complex does not work in NHEJ. Instead, this role is taken up by DNA-PK. Human homologs of the different yeast proteins are listed in the table.
NHEJ is initiated by binding of the Ku protein, a heterodimer composed of the subunits Yku70 and Yku80, and the MRX complex to the DNA ends of the DSB [190]. This binding blocks resection and prevents formation of long ssDNA tracts necessary for initiation of HR [191,192]. The role of the MRX complex, which also functions in HR, is not well understood. It is thought that MRX functions as a complex that bridges DSB ends and acts as a flexible tether to assist ligation in NHEJ [193]. Tethering and protection of the ends stabilizes the ends and brings them together. The Ku complex then recruits other core NHEJ proteins, such as Dnl4-Lif1 and Nej1, to the DSB site forming the repair super complex. End-processing factors such as Pol4 act in parallel with the complex formation and are required to make complementary ends needed for ligation. Pol4 is known to initiate gap filling at unstable 3′ overhangs [194,195]. After end-processing ligation is carried out by DNA ligase IV, Dnl4 in S. cerevisiae, stabilized by Lif1 [196,197]. To ensure stable formation of the NHEJ super complex at DSB ends, Nej1 functions as a stabilizing factor [198].
Even though the process of NHEJ is largely similar between yeast and humans, the initiation of NHEJ is performed by different proteins. In humans, Ku70/Ku80 heterodimers recruit DNA-dependent protein kinase catalytic subunit (DNA-Pkc) which undergoes auto-phosphorylation and thereby recruits other repair factors [199]. Next, end processing is performed by the Artemis endonuclease [200]. For both DNA-Pkc and Artemis, no homologous have been identified in S. cerevisiae.
Repair pathways for different types of base damages, such as BER, NER, and MMR rely on specialized enzymes for the recognition of the type of damage. However, the repair of DSBs can be performed by both HR and NHEJ, and the choice of which DSB repair pathway is activated depends on regulatory processes that are influenced by the cell type, the structure of the DNA ends resulting from resection, as well as the phase of the cell cycle. In both yeast and humans, HR is most active in the S and G2 phase when homologous templates are available. NHEJ on the other hand is active during the complete cell cycle [191,201,202]. This indicates that HR and NHEJ compete at DSBs. The choice between HR and NHEJ is then controlled by the extent of 5′–3′ end resection determined by the Ku heterodimer, the MRX complex, and the endonuclease Sae2. HR starts with extensive end resection of the 5′ strand to generate ssDNA which is bound by RPA. Once resection has started, the long ssDNA ends become poor substrates for Yku70 and Yku80 [203].
Since yeast is less efficient in NHEJ, NHEJ was only observed in strains lacking genes involved in HR [204]. Moreover, deletion of yeast NHEJ genes in the presence of functional HR genes does not result in sensitivity to several DNA damaging agents. This makes studying NHEJ in yeast more difficult using conventional molecular and genetic assays. Nonetheless, elegantly designed assays can overcome this problem. Ooi et al. for example, used an in vivo plasmid repair assay to screen the yeast deletion collection for genes involved in NHEJ. In their assay, they transformed the yeast deletion collection with circular and linearized plasmids and measured the transformation efficiency as a quantitative readout. Deletion mutants with an active NHEJ process will be able to repair the linearized plasmid and grow to similar levels as when they are transformed with a circular plasmid. Deletion mutants with a defect in NHEJ fail to recircularize the plasmid which will result in a lower transformation efficiency. Using this assay, they were able to identify the gene NEJ1 [205].
3. Challenges and Future Perspectives of Yeast as a Model System
Multiple DDR kinases, such as Dun1, Mec1, Chk1, and Rad53, were first identified and characterized in yeast [44,101,206,207,208]. Additionally, genome-wide screens of the yeast deletion collection with DNA damage inducers such as MMS, UV light or ionizing radiation further continued to identify genes involved in the DNA damage response [93,209,210]. Many of the genes identified in yeast proved to be conserved in higher eukaryotes, and as such, the yeast work much facilitated the unraveling of DNA damage response mechanisms in other organisms. However, using yeast as a model organism for higher, more complex organisms does come with some pitfalls and thus results cannot always be translated without verification. Most studies in yeast are performed in haploids even though more complex organisms such as humans are diploid. Haploid genomes are easier to manipulate and make it easier to see the effect of certain mutations, deletions, etc. However, diploids and haploids can show a different regulation of certain DNA repair pathways. For example, diploids yeast cells are known to exhibit lower levels of NHEJ than haploid cells [211,212,213,214].
Moreover, pathways for double-strand break repair are differentially favored in yeast compared to humans. Early experiments in yeast, where cell killing by radiation was investigated, seemed to indicate that HR was the most dominant DSB repair mechanism in yeast. The type of DNA ends produced by radiation do not form a suitable template for NHEJ as they first require processing by the MRX complex. In mammals on the other hand, NHEJ is the preferred pathway for repair of DSBs. It has been observed that the process of NHEJ is faster in than HR in humans [201]. Moreover, due to the highly repetitive nature of human genomes, efficient HR could lead to deleterious genomic rearrangements.
Why yeast NHEJ is so inefficient is unclear. The most likely reason is that mammals have more NHEJ proteins for which no homolog is present in yeast [215]. Although S. cerevisiae possesses all core NHEJ factors, it lacks the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) that in mammals is recruited to DSBs by the Ku complex and is required for NHEJ. As a result, the mammalian DNA damage response is coordinated by three DNA damage sensors: ATM, ATR, and DNA-PK. In yeast, the MRX complex compensates for the lack of DNA-PK, but perhaps this does result in lower NHEJ activity [190,193]. This difference between NHEJ in yeast and human illustrates the more complex nature of the mammalian DNA damage response. Although the core processes of the DNA damage response are largely similar, in mammals, much more genes are involved in the DNA damage response. To elucidate the functions of all these genes, more research is required.
Since the toolbox for studying higher eukaryotes steadily expands, including, for example, the implementation of various technologies based on CRISPR-Cas, the need to use a model system may appear to decline. However, the yeast continues to be an important model system to further unravel the full functioning and coordination of the DNA damage response. It has become clear that DNA damage response kinases also regulate other processes such as carbon metabolism, autophagy, and protein homeostasis [216,217,218,219]. Moreover, the list of genes that are shown to play a role in the DNA damage response is still steadily increasing [220]. For example, the recent focus on phosphoproteomics signaling identified new targets of the DNA damage kinases [221,222]. Moreover, the exact molecular function of specific DNA damage response proteins is also being investigated in yeast [223,224].
These new studies in the field of DNA damage response also result in the development of multiple new techniques. For example, in order to study the human DNA repair proteins using a yeast system, humanized yeasts form an excellent tool. Many protein-coding human genes can successfully substitute for their yeast equivalents and sustain yeast growth [36,225,226]. This allows us to study the function of human proteins in a less complex organism and to characterize several cancer-associated mutations and their influence on protein functioning [227,228,229].
In addition, Duan et al. studied the genome-wide role of Rad26, the protein that initiates TC-NER, using chromatin immunoprecipitation sequencing (ChIP-seq) [230]. Peritore et al. used strand-specific ChIP-seq to determine which DNA repair proteins associated with the ssDNA and dsDNA, respectively, at a DSB site. They were able to confirm the ssDNA-binding nature of RPA and Rad51 but also visualized the dsDNA-binding nature of Rad51 during the homology search. Moreover, they showed that nucleosomes are removed during the resection process and are thus not associated with ssDNA at the site of damage [231].
Alternative versions of the inducible HO-system have also been developed. One of the shortcomings of the galactose inducible HO-system is that it requires specific nutrient conditions for its induction. This limits the metabolic states in which DSB signaling and repair can be studied. Moreover, efficient galactose-induction requires cells to be precultured in a non-fermentable carbon source. This can become tedious in the case of DSB repair mutant strains since they often have a defect in cell growth. To overcome these limitations, recently a media-independent heterologous induction system that controls HO expression was developed. Using β-estradiol they were able to induce a single DSB in an efficient manner under different carbon sources such as glucose, lactic acid and galactose [232]. Additionally, using several restriction enzymes that specifically recognize between 20 and 100 8 bp recognition sites in the yeast genome, they were able to develop an inducible system that results in multiple DSBs at defined loci [232].
Technologies based on CRISPR-Cas have emerged as a powerful genome editing tool. The outcome of the CRISPR-Cas-induced double-strand break can vary depending on the repair pathway that is used. While NHEJ results in insertions or deletions at the targeted locus, HR results in precise mutations or complete repair by using the homologous sequence that is provided. Vyas et al. showed that different yeasts rely differentially on either HR or NHEJ for the repair of the CRISPR-Cas9-induced DSB. While S. cerevisiae and Candida albicans rely on HR, Candida glabrata and Naumovozyma castellii mainly rely on NHEJ [233]. Consequently, because different organisms have different primary repair pathways, specific genetic editing needs need to be taken into account in order to optimize CRISPR-Cas9 mutagenesis. Lemos et al. studied how binding of the Cas9::gRNA complex influenced DSB repair by NHEJ. Therefore, they assessed the repair profiles of gRNAs pairs that were complementary to opposite DNA strands and cleaved at the same chromosomal location. They repeatedly found insertions of 1 base, in which the added base was dependent on the orientation of Cas9::gRNA binding [234]. Studying the repair of CRISPR-Cas induced DSBs in yeast can further improve the development of this genome editing tool.
In addition, in radiation biology, yeast served as an important model organism to study the response to radiation. As previously mentioned, Hartwell introduced his concept of checkpoints based on observations made after radiation treatment [40]. In addition, multiple DNA damage response genes have been identified using radiation to induce DNA damage; hence, many of these genes are termed “rad”-genes [206,235,236]. Even now, with the increased toolbox for higher eukaryotes, yeast remains a useful model in radiation biology. For example, to investigate radiation damage and repair after different radiation modalities, such as conventional photon radiotherapy and proton radiotherapy [15,237].
Together, it is clear that despite the shortcomings and pitfalls, yeast remains an important model system to further unravel the molecular mechanisms of proteins functioning in the DNA damage response, as well as to further explore the coordination and regulation of the DNA damage response. As such, the simplicity of the yeast model remains key in unraveling and understanding a complex pathway as the DNA damage response.
4. Conclusions
Besides its indispensable role in food production, the yeast S. cerevisiae proved an excellent model organism to study basic eukaryotic biology, as exemplified by the long list of Nobel prizes that have been awarded to yeast researchers. The existing molecular toolbox, together with the extensive genetic and physiological knowledge and the high conservation of core processes, make S. cerevisiae a particularly suitable and popular model for to study of eukaryotic biology. New techniques and approaches for studying biological processes, such as different omics approaches, keep being developed and continue to increase our knowledge.
The DNA damage response is a prime example of a eukaryotic pathway in which the key steps were first unraveled using yeast as a model organism. Especially HR was extensively studied in yeast using the HO locus to introduce one single defined double-strand break. In addition, genome-wide screens using the yeast deletion collection identified multiple genes involved in the DNA damage response to different DNA damage inducers.
It should be noted, however, that using yeast as a model for more complex organisms comes with pitfalls. However, the yeast model remains key in unraveling and understanding complex pathways, as it seems that new technologies and findings often still first emerge in this fantastic organism.
Author Contributions
L.V., R.D., K.V., S.N. and K.J.V. contributed to the research, drafting, and editing of this manuscript. L.V., R.D., K.V., S.N. and K.J.V. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the KU Leuven Research Fund (C24/17/073). R.D. is supported by the Emmanuel van der Schueren fellowship for postdoctoral researchers from Kom op Tegen Kanker and by the Flemish Foundation of Scientific Research (FWO-Vlaanderen, 1521019N). S.N. is supported by a clinical research mandate from the Flemish Foundation of Scientific Research (FWO-Vlaanderen, 18B4122N). Research in the laboratory of K.J.V. is supported by VIB, KU Leuven, FWO, and European Research Council (ERC) Consolidator Grant CoG682009.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Liu L., Wang J., Rosenberg D., Zhao H., Lengyel G., Nadel D. Fermented beverage and food storage in 13,000 y-old stone mortars at Raqefet Cave, Israel: Investigating Natufian ritual feasting. J. Archaeol. Sci. Rep. 2018;21:783–793. doi: 10.1016/j.jasrep.2018.08.008. [DOI] [Google Scholar]
- 2.Arranz-Otaegui A., Carretero L.G., Ramsey M.N., Fuller D.Q., Richter T. Archaeobotanical evidence reveals the origins of bread 14,400 years ago in northeastern Jordan. Proc. Natl. Acad. Sci. USA. 2018;115:7925–7930. doi: 10.1073/pnas.1801071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasteur L. Études Sur la Bière. Gauthier-Villars; Paris, France: 1876. [Google Scholar]
- 4.Maicas S. The Role of Yeasts in Fermentation Processes. Microorganisms. 2020;8:1142. doi: 10.3390/microorganisms8081142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glamann K., Glamann K., Carlsbergfondet . The Story of Emil Chr. Hansen. Carlsberg Foundation; Valby, Copenhagen: 2009. [Google Scholar]
- 6.Hinnen A., Hicks J.B., Fink G.R. Transformation of yeast. Genetics. 1978;75:1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goffeau A., Barrell G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., et al. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 8.Peter J., De Chiara M., Friedrich A., Yue J.X., Pflieger D., Bergström A., Sigwalt A., Barre B., Freel K., Llored A., et al. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature. 2018;556:339–344. doi: 10.1038/s41586-018-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Y.O., Sherlock G., Petrov D.A. Whole genome analysis of 132 clinical Saccharomyces cerevisiae strains reveals extensive ploidy variation. G3 Genes Genomes Genet. 2016;6:2421–2434. doi: 10.1534/G3.116.029397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallone B., Steensels J., Prahl T., Soriaga L., Saels V., Herrera-Malaver B., Merlevede A., Roncoroni M., Voordeckers K., Miraglia L., et al. Domestication and Divergence of Saccharomyces cerevisiae Beer Yeasts. Cell. 2016;166:1397–1410.e16. doi: 10.1016/j.cell.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 12.Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 13.Dowell R.D., Ryan O., Jansen A., Cheung D., Agarwala S., Danford T., Bernstein D.A., Alexander Rolfe P., Heisler L.E., Chin B., et al. Genotype to phenotype: A Complex problem. Science. 2010;328:469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Birrell G.W., Giaever G., Chu A.M., Davis R.W., Brown J.M. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc. Natl. Acad. Sci. USA. 2001;98:12608–12613. doi: 10.1073/pnas.231366398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vanderwaeren L., Dok R., Voordeckers K., Vandemaele L., Verstrepen K.J., Nuyts S. An Integrated Approach Reveals DNA Damage and Proteotoxic Stress as Main Effects of Proton Radiation in S. cerevisiae. Int. J. Mol. Sci. 2022;23:5493. doi: 10.3390/ijms23105493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helsen J., Voordeckers K., Vanderwaeren L., Santermans T., Tsontaki M., Verstrepen K.J., Jelier R. Gene loss predictably drives evolutionary adaptation. Mol. Biol. Evol. 2020;37:2989–3002. doi: 10.1093/molbev/msaa172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Samper G., Cerulus B., Jariani A., Vermeersch L., Simancas N.B., Bisschops M.M.M., van den Brink J., Solis-Escalante D., Gallone B., De Maeyer D., et al. The crabtree effect shapes the Saccharomyces cerevisiae lag phase during the switch between different carbon sources. MBio. 2018;9:e01331-18. doi: 10.1128/mBio.01331-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun S., Baryshnikova A., Brandt N., Gresham D. Genetic interaction profiles of regulatory kinases differ between environmental conditions and cellular states. Mol. Syst. Biol. 2020;16:e9167. doi: 10.15252/msb.20199167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berns K., Hijmans E.M., Mullenders J., Brummelkamp T.R., Velds A., Heimerikx M., Kerkhoven R.M., Madlredjo M., Nijkamp W., Weigelt B., et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 20.Brummelkamp T.R., Bernards R. New tools for functional mammalian cancer genetics. Nat. Rev. Cancer. 2003;3:781–789. doi: 10.1038/nrc1191. [DOI] [PubMed] [Google Scholar]
- 21.Bock C., Datlinger P., Chardon F., Coelho M.A., Dong M.B., Lawson K.A., Lu T., Maroc L., Norman T.M., Song B., et al. High-content CRISPR screening. Nat. Rev. Methods Prim. 2022;2:8. doi: 10.1038/s43586-021-00093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 24.Jones G.M., Stalker J., Humphray S., West A., Cox T., Rogers J., Dunham I., Prelich G. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nat. Methods. 2008;5:239–241. doi: 10.1038/nmeth.1181. [DOI] [PubMed] [Google Scholar]
- 25.DeRisi J.L., Iyer V.R., Brown P.O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 26.Lashkari D.A., Derisi J.L., Mccusker J.H., Namath A.F., Gentile C., Hwang S.Y., Brown P.O., Davis R.W. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc. Natl. Acad. Sci. USA. 1997;94:13057. doi: 10.1073/pnas.94.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 28.Villas-Bôas S.G., Moxley J.F., Åkesson M., Stephanopoulos G., Nielsen J. High-throughput metabolic state analysis: The missing link in integrated functional genomics of yeasts. Biochem. J. 2005;388:669. doi: 10.1042/BJ20041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jewett M.C., Hofmann G., Nielsen J. Fungal metabolite analysis in genomics and phenomics. Curr. Opin. Biotechnol. 2006;17:191–197. doi: 10.1016/j.copbio.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Picotti P., Clément-Ziza M., Lam H., Campbell D.S., Schmidt A., Deutsch E.W., Röst H., Sun Z., Rinner O., Reiter L., et al. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature. 2013;494:266–270. doi: 10.1038/nature11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson D.S., Mortazavi A., Myers R.M., Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 32.Saccharomyces Genome Database. [(accessed on 16 March 2022)]. Available online: https://www.yeastgenome.org/
- 33.Cherry J.M., Hong E.L., Amundsen C., Balakrishnan R., Binkley G., Chan E.T., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., et al. Saccharomyces Genome Database: The genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee M.G., Nurse P. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature. 1987;327:31–35. doi: 10.1038/327031a0. [DOI] [PubMed] [Google Scholar]
- 35.Douzery E.J.P., Snell E.A., Bapteste E., Delsuc F., Philippe H. The timing of eukaryotic evolution: Does a relaxed molecular clock reconcile proteins and fossils? Proc. Natl. Acad. Sci. USA. 2004;101:15386–15391. doi: 10.1073/pnas.0403984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kachroo A.H., Laurent J.M., Yellman C.M., Meyer A.G., Wilke C.O., Marcotte E.M. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science. 2015;348:921–925. doi: 10.1126/science.aaa0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Culotti J., Hartwell L.H. Genetic control of the cell division cycle in yeast. III. Seven genes controlling nuclear division. Exp. Cell Res. 1971;67:389–401. doi: 10.1016/0014-4827(71)90424-1. [DOI] [PubMed] [Google Scholar]
- 38.Hartwell L.H. Genetic control of the cell division cycle in yeast. II. Genes controlling DNA replication and its initiation. J. Mol. Biol. 1971;59:183–194. doi: 10.1016/0022-2836(71)90420-7. [DOI] [PubMed] [Google Scholar]
- 39.Hartwell L.H. Genetic control of the cell division cycle in yeast: IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- 40.Siede W., Friedberg A.S., Friedberg E.C. RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1993;90:7985–7989. doi: 10.1073/pnas.90.17.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartwell L.H., Weinert T.A. Checkpoints: Controls That Ensure the Order of Cell Cycle Events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 42.Paulovich A.G., Toczyski D.P., Hartwell L.H. When Checkpoints Fail. Cell. 1997;88:315–321. doi: 10.1016/S0092-8674(00)81870-X. [DOI] [PubMed] [Google Scholar]
- 43.Weinert T.A., Hartwell L.H. Cell Cycle Arrest of Cdc Mutants and Specificity of the Rad9 Checkpoint. Genetics. 1993;134:63. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinert T.A., Kiser G.L., Hartwell L.H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 45.Thuriaux P., Nurse P., Carter B. Mutants altered in the control co-ordinating cell division with cell growth in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 1978;161:215–220. doi: 10.1007/BF00274190. [DOI] [PubMed] [Google Scholar]
- 46.Nurse P., Thuriaux P. Regulatory genes controlling mitosis in the fission yeast Schizosaccharomyces pombe. Genetics. 1980;96:627–637. doi: 10.1093/genetics/96.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nasmyth K., Nurse P. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 1981;182:119–124. doi: 10.1007/BF00422777. [DOI] [PubMed] [Google Scholar]
- 48.Nurse P., Bissett Y. Gene required in G1 for commitment to cell cycle and in G2 for control of mitosis in fission yeast. Nature. 1981;292:558–560. doi: 10.1038/292558a0. [DOI] [PubMed] [Google Scholar]
- 49.Forsburg S.L., Nurse P. Cell cycle regulation in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Annu. Rev. Cell Biol. 1991;7:227–256. doi: 10.1146/annurev.cb.07.110191.001303. [DOI] [PubMed] [Google Scholar]
- 50.Hartwell L. Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell. 1992;71:543–546. doi: 10.1016/0092-8674(92)90586-2. [DOI] [PubMed] [Google Scholar]
- 51.Hartwell L.H., Kastan M.B. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 52.Hartwell L.H. Nobel Lecture. Yeast and cancer. Biosci. Rep. 2002;22:373–394. doi: 10.1023/A:1020918107706. [DOI] [PubMed] [Google Scholar]
- 53.Kornberg R.D. Chromatin structure: A repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 54.Kornberg R.D., Thomas J.O. Chromatin structure; oligomers of the histones. Science. 1974;184:865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- 55.Lue N.F., Kornberg R.D. Accurate initiation at RNA polymerase II promoters in extracts from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1987;84:8839. doi: 10.1073/pnas.84.24.8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sayre M.H., Tschochner H., Kornberg R.D. Reconstitution of transcription with five purified initiation factors and RNA polymerase II from Saccharomyces cerevisiae. J. Biol. Chem. 1992;267:23376–23382. doi: 10.1016/S0021-9258(18)50101-0. [DOI] [PubMed] [Google Scholar]
- 57.Chasman D.I., Flaherty K.M., Sharp P.A., Kornberg R.D. Crystal structure of yeast TATA-binding protein and model for interaction with DNA. Proc. Natl. Acad. Sci. USA. 1993;90:8174–8178. doi: 10.1073/pnas.90.17.8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Asturias F.J., Jiang Y.W., Myers L.C., Gustafsson C.M., Kornberg R.D. Conserved structures of mediator and RNA polymerase II holoenzyme. Science. 1999;283:985–987. doi: 10.1126/science.283.5404.985. [DOI] [PubMed] [Google Scholar]
- 59.Gnatt A.L., Cramer P., Fu J., Bushnell D.A., Kornberg R.D. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 Å resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 60.Bushnell D.A., Cramer P., Kornberg R.D. Structural basis of transcription: α-Amanitin–RNA polymerase II cocrystal at 2.8 Å resolution. Proc. Natl. Acad. Sci. USA. 2002;99:1218. doi: 10.1073/pnas.251664698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szostak J.W., Blackburn E.H. Cloning yeast telomeres on linear plasmid vectors. Cell. 1982;29:245–255. doi: 10.1016/0092-8674(82)90109-X. [DOI] [PubMed] [Google Scholar]
- 62.Murray A.W., Szostak J.W. Construction of artificial chromosomes in yeast. Nature. 1983;305:189–193. doi: 10.1038/305189a0. [DOI] [PubMed] [Google Scholar]
- 63.Shampay J., Szostak J.W., Blackburn E.H. DNA sequences of telomeres maintained in yeast. Nature. 1984;310:154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- 64.Lundblad V., Szostak J.W. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 65.Blackburn E.H., Gall J.G. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J. Mol. Biol. 1978;120:33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- 66.Greider C.W., Blackburn E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 67.Greider C.W., Blackburn E.H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 68.Yu G.L., Bradley J.D., Attardi L.D., Blackburn E.H. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature. 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- 69.Mc Eachern M.J., Blackburn E.H. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 70.Kim M.M., Rivera M.A., Botchkina I.L., Shalaby R., Thor A.D., Blackburn E.H. A low threshold level of expression of mutant-template telomerase RNA inhibits human tumor cell proliferation. Proc. Natl. Acad. Sci. USA. 2001;98:7982–7987. doi: 10.1073/pnas.131211098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Novick P., Schekman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1979;76:1858. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Novick P., Field C., Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 73.Deshaies R.J., Schekman R. A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J. Cell Biol. 1987;105:633. doi: 10.1083/jcb.105.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baker D., Hicke L., Rexach M., Schleyer M., Schekman R. Reconstitution of SEC gene product-dependent intercompartmental protein transport. Cell. 1988;54:335–344. doi: 10.1016/0092-8674(88)90196-1. [DOI] [PubMed] [Google Scholar]
- 75.Barlowe C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N., Rexach M.F., Ravazzola M., Amherdt M., Schekman R. COPII: A membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 76.Matsuoka K., Orci L., Amherdt M., Bednarek S.Y., Hamamoto S., Schekman R., Yeung T. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/S0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- 77.Kaiser C.A., Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-U. [DOI] [PubMed] [Google Scholar]
- 78.Balch W.E., Dunphy W.G., Braell W.A., Rothman J.E. Reconstitution of the transport of protein between successive compartments of the Golgi measured by the coupled incorporation of N-acetylglucosamine. Cell. 1984;39:405–416. doi: 10.1016/0092-8674(84)90019-9. [DOI] [PubMed] [Google Scholar]
- 79.Söllner T., Whiteheart S.W., Brunner M., Erdjument-Bromage H., Geromanos S., Tempst P., Rothman J.E. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 80.Tsukada M., Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-E. [DOI] [PubMed] [Google Scholar]
- 81.Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1992;119:301–312. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M.D., Klionsky D.J., Ohsumi M., Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 83.Foury F. Human genetic diseases: A cross-talk between man and yeast. Gene. 1997;195:1–10. doi: 10.1016/S0378-1119(97)00140-6. [DOI] [PubMed] [Google Scholar]
- 84.Mager W.H., Winderickx J. Yeast as a model for medical and medicinal research. Trends Pharmacol. Sci. 2005;26:265–273. doi: 10.1016/j.tips.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 85.Giaever G., Shoemaker D.D., Jones T.W., Liang H., Winzeler E.A., Astromoff A., Davis R.W. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat. Genet. 1999;21:278–283. doi: 10.1038/6791. [DOI] [PubMed] [Google Scholar]
- 86.Lum P.Y., Armour C.D., Stepaniants S.B., Cavet G., Wolf M.K., Butler J.S., Hinshaw J.C., Garnier P., Prestwich G.D., Leonardson A., et al. Discovering Modes of Action for Therapeutic Compounds Using a Genome-Wide Screen of Yeast Heterozygotes. Cell. 2004;116:121–137. doi: 10.1016/S0092-8674(03)01035-3. [DOI] [PubMed] [Google Scholar]
- 87.Giaever G., Flaherty P., Kumm J., Proctor M., Nislow C., Jaramillo D.F., Chu A.M., Jordan M.I., Arkin A.P., Davis R.W. Chemogenomic profiling: Identifying the functional interactions of small molecules in yeast. Proc. Natl. Acad. Sci. USA. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bivi N., Romanello M., Harrison R., Clarke I., Hoyle D.C., Moro L., Ortolani F., Bonetti A., Quadrifoglio F., Tell G., et al. Identification of secondary targets of N-containing bisphosphonates in mammalian cells via parallel competition analysis of the barcoded yeast deletion collection. Genome Biol. 2009;10:R93. doi: 10.1186/gb-2009-10-9-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chan T.F., Carvalho J., Riles L., Zheng X.F.S. A chemical genomics approach toward understanding the global functions of the target of rapamycin protein (TOR) Proc. Natl. Acad. Sci. USA. 2000;97:13227–13232. doi: 10.1073/pnas.240444197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zewail A., Xie M.W., Xing Y., Lin L., Fred Zhang P., Zou W., Saxe J.P., Huang J. Novel functions of the phosphatidylinositol metabolic pathway discovered by a chemical genomics screen with wortmannin. Proc. Natl. Acad. Sci. USA. 2003;100:3345–3350. doi: 10.1073/pnas.0530118100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Parsons A.B., Lopez A., Givoni I.E., Williams D.E., Gray C.A., Porter J., Chua G., Sopko R., Brost R.L., Ho C.-H., et al. Exploring the Mode-of-Action of Bioactive Compounds by Chemical-Genetic Profiling in Yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 92.Parsons A.B., Brost R.L., Ding H., Li Z., Zhang C., Sheikh B., Brown G.W., Kane P.M., Hughes T.R., Boone C. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 93.Game J.C., Birrell G.W., Brown J.A., Shibata T., Baccari C., Chu A.M., Williamson M.S., Brown J.M. Use of a genome-wide approach to identify new genes that control resistance of Saccharomyces cerevisiae to ionizing radiation. Radiat. Res. 2003;160:14–24. doi: 10.1667/RR3019. [DOI] [PubMed] [Google Scholar]
- 94.Nakada D., Matsumoto K., Sugimoto K. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 2003;17:1957–1962. doi: 10.1101/gad.1099003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Grenon M., Gilbert C., Lowndes N.F. Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat. Cell Biol. 2001;3:844–847. doi: 10.1038/ncb0901-844. [DOI] [PubMed] [Google Scholar]
- 96.Hustedt N., Gasser S.M., Shimada K. Replication checkpoint: Tuning and coordination of replication forks in S phase. Genes. 2013;4:388–434. doi: 10.3390/genes4030388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smith J., Mun Tho L., Xu N., Gillespie D.A. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 98.Lanz M.C., Dibitetto D., Smolka M.B. DNA damage kinase signaling: Checkpoint and repair at 30 years. EMBO J. 2019;38:e101801. doi: 10.15252/embj.2019101801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao X., Rothstein R. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc. Natl. Acad. Sci. USA. 2002;99:3746–3751. doi: 10.1073/pnas.062502299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gasch A.P., Huang M., Metzner S., Botstein D., Elledge S.J., Brown P.O. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR Homolog Mec1p. Mol. Biol. Cell. 2001;12:2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou Z., Elledge S.J. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell. 1993;75:1119–1127. doi: 10.1016/0092-8674(93)90321-G. [DOI] [PubMed] [Google Scholar]
- 102.Allen J.B., Zhou Z., Siede W., Friedberg E.C., Elledge S.J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 103.Tanaka H., Arakawa H., Yamaguchi T., Shiraishi K., Fukuda S., Matsui K., Takei Y., Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 104.Beard W.A., Horton J.K., Prasad R., Wilson S.H. Eukaryotic Base Excision Repair: New Approaches Shine Light on Mechanism. Annu. Rev. Biochem. 2019;88:137–162. doi: 10.1146/annurev-biochem-013118-111315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Van Der Kemp P.A., Thomas D., Barbey R., De Oliveira R., Boiteux S. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc. Natl. Acad. Sci. USA. 1996;93:5197–5202. doi: 10.1073/pnas.93.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Eide L., Bjørås M., Pirovano M., Alseth I., Berdal K.G., Seeberg E. Base excision of oxidative purine and pyrimidine DNA damage in Saccharomyces cerevisiae by a DNA glycosylase with sequence similarity to endonuclease III from Escherichia coli. Proc. Natl. Acad. Sci. USA. 1996;93:10735–10740. doi: 10.1073/pnas.93.20.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Crosby B., Prakash L., Davis H., Hinkle D.C. Purification and characterization of a uracil-DNA glycosylase from the yeast. Saccharomyces cerevisiae. Nucleic Acids Res. 1981;9:5797. doi: 10.1093/nar/9.21.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Prakash L., Prakash S. Isolation and Characterization of Mms-Sensitive Mutants of Saccharomyces cerevisiae. Genetics. 1977;86:33. doi: 10.1093/genetics/86.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Samanta M.P., Liang S. Predicting protein functions from redundancies in large-scale protein interaction networks. Proc. Natl. Acad. Sci. USA. 2003;100:12579. doi: 10.1073/pnas.2132527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huffman J.L., Sundheim O., Tainer J.A. DNA base damage recognition and removal: New twists and grooves. Mutat. Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 111.Hindi N.N., Elsakrmy N., Ramotar D. The base excision repair process: Comparison between higher and lower eukaryotes. Cell. Mol. Life Sci. 2021;78:7943–7965. doi: 10.1007/s00018-021-03990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Johnson R.E., Torres-Ramos C.A., Izumi T., Mitra S., Prakash S., Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 1998;12:3137. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Popoff B.C., Spira A.I., Johnson A.W., Demple B. Yeast structural gene (APN1) for the major apurinic endonuclease: Homology to Escherichia coli endonuclease IV. Proc. Natl. Acad. Sci. USA. 1990;87:4193–4197. doi: 10.1073/pnas.87.11.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Prasad R., Batra V.K., Yang X.P., Krahn J.M., Pedersen L.C., Beard W.A., Wilson S.H. Structural insight into the DNA polymerase β deoxyribose phosphate lyase mechanism. DNA Repair. 2005;4:1347–1357. doi: 10.1016/j.dnarep.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 115.Svilar D., Goellner E.M., Almeida K.H., Sobol R.W. Base excision repair and lesion-dependent subpathways for repair of oxidative DNA damage. Antioxid. Redox Signal. 2011;14:2491–2507. doi: 10.1089/ars.2010.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Reagan M.S., Pittenger C., Siede W., Friedberg E.C. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 1995;177:364. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Citarelli M., Teotia S., Lamb R.S. Evolutionary history of the poly(ADP-ribose) polymerase gene family in eukaryotes. BMC Evol. Biol. 2010;10:308. doi: 10.1186/1471-2148-10-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Arai K., Morishita K., Shinmura K., Kohno T., Kim S.R., Nohmi T., Taniwaki M., Ohwada S., Yokota J. Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene. 1997;14:2857–2861. doi: 10.1038/sj.onc.1201139. [DOI] [PubMed] [Google Scholar]
- 119.Rosenquist T.A., Zharkov D.O., Grollman A.P. Cloning and characterization of a mammalian 8-oxoguanine dna glycosylase. Proc. Natl. Acad. Sci. USA. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Aburatani H., Hippo Y., Ishida T., Takashima R., Matsuba C., Kodama T., Takao M., Yasui A., Yamamoto K., Asano M., et al. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- 121.Lavrik O.I., Prasad R., Sobol R.W., Horton J.K., Ackerman E.J., Wilson S.H. Photoaffinity labeling of mouse fibroblast enzymes by a base excision repair intermediate. Evidence for the role of poly(ADP-ribose) polymerase-1 in DNA repair. J. Biol. Chem. 2001;276:25541–25548. doi: 10.1074/jbc.M102125200. [DOI] [PubMed] [Google Scholar]
- 122.Cabrera M., Nghiem Y., Miller J.H. mutM, a second mutator locus in Escherichia coli that generates G.C----T.A transversions. J. Bacteriol. 1988;170:5405–5407. doi: 10.1128/jb.170.11.5405-5407.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ganai R.A., Johansson E. Molecular Cell DNA Replication-A Matter of Fidelity. Mol. Cell. 2016;62:745–755. doi: 10.1016/j.molcel.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 124.Lynch M., Sung W., Morris K., Coffey N., Landry C.R., Dopman E.B., Dickinson W.J., Okamoto K., Kulkarni S., Hartl D.L., et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. USA. 2008;105:9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Modrich P. Mismatch Repair in Replication Fidelity, Genetic Recombination, and Cancer Biology. Annu. Rev. Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 126.Shin-San S., Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc. Natl. Acad. Sci. USA. 1986;83:5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Acharya S., Wilson T., Gradia S., Kane M.F., Guerrette S., Marsischky G.T., Kolodner R., Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc. Natl. Acad. Sci. USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hughes M.J., Jiricny J. The purification of a human mismatch-binding protein and identification of its associated ATPase and helicase activities. J. Biol. Chem. 1992;267:23876–23882. doi: 10.1016/S0021-9258(18)35918-0. [DOI] [PubMed] [Google Scholar]
- 129.Buermeyer A.B., Deschênes S.M., Baker S.M., Liskay R.M. Mammalian DNA Mismatch Repair. Annu. Rev. Genet. 1999;33:533–564. doi: 10.1146/annurev.genet.33.1.533. [DOI] [PubMed] [Google Scholar]
- 130.Tishkoff D.X., Boerger A.L., Bertrand P., Filosi N., Gaida G.M., Kane M.F., Kolodner R.D. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc. Natl. Acad. Sci. USA. 1997;94:7487. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tishkoff D.X., Amin N.S., Viars C.S., Arden K.C., Kolodner R.D. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res. 1998;58:5027–5031. [PubMed] [Google Scholar]
- 132.Schmutte C., Marinescu R.C., Sadoff M.M., Guerrette S., Overhauser J., Fishel R. Human exonuclease I interacts with the mismatch repair protein hMSH2. Cancer Res. 1998;58:4537–4542. [PubMed] [Google Scholar]
- 133.Alani E., Sokolsky T., Studamire B., Miret J.J., Lahue R.S. Genetic and biochemical analysis of Msh2p-Msh6p: Role of ATP hydrolysis and Msh2p-Msh6p subunit interactions in mismatch base pair recognition. Mol. Cell. Biol. 1997;17:2436–2447. doi: 10.1128/MCB.17.5.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Blackwell L.J., Martik D., Bjornson K.P., Bjornson E.S., Modrich P. Nucleotide-promoted release of hMutSα from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. J. Biol. Chem. 1999;273:32055–32062. doi: 10.1074/jbc.273.48.32055. [DOI] [PubMed] [Google Scholar]
- 135.Gradia S., Subramanian D., Wilson T., Acharya S., Makhov A., Griffith J., Fishel R. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell. 1999;3:255–261. doi: 10.1016/S1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- 136.Ban C., Junop M., Yang W. Transformation of MutL by ATP binding and hydrolysis: A switch in DNA mismatch repair. Cell. 1999;97:85–97. doi: 10.1016/S0092-8674(00)80717-5. [DOI] [PubMed] [Google Scholar]
- 137.Flores-Rozas H., Clark D., Kolodner R.D. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat. Genet. 2000;26:375–378. doi: 10.1038/81708. [DOI] [PubMed] [Google Scholar]
- 138.Johnson R.E., Kovvali G.K., Guzder S.N., Amin N.S., Holm C., Habraken Y., Sung P., Prakash L., Prakash S. Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. J. Biol. Chem. 1996;271:27987–27990. doi: 10.1074/jbc.271.45.27987. [DOI] [PubMed] [Google Scholar]
- 139.Umar A., Buermeyer A.B., Simon J.A., Thomas D.C., Clark A.B., Liskay R.M., Kunkel T.A. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/S0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 140.Constantin N., Dzantiev L., Kadyrov F.A., Modrich P. Human mismatch repair: Reconstitution of a nick-directed bidirectional reaction. J. Biol. Chem. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang Y., Yuan F., Presnell S.R., Tian K., Gao Y., Tomkinson A.E., Gu L., Li G.M. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 142.Guzder S.N., Sung P., Prakash L., Prakash S. Affinity of yeast nucleotide excision repair factor 2, consisting of the Rad4 and Rad23 proteins, for ultraviolet damaged DNA. J. Biol. Chem. 1998;273:31541–31546. doi: 10.1074/jbc.273.47.31541. [DOI] [PubMed] [Google Scholar]
- 143.Van Gool A.J., Verhage R., Swagemakers S.M.A., Van De Putte P., Brouwer J., Troelstra C., Bootsma D., Hoeijmakers J.H.J. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 1994;13:5361. doi: 10.1002/j.1460-2075.1994.tb06871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bhatia P.K., Verhage R.A., Brouwer J., Friedberg E.C. Molecular cloning and characterization of Saccharomyces cerevisiae RAD28, the yeast homolog of the human Cockayne syndrome A (CSA) gene. J. Bacteriol. 1996;178:5977. doi: 10.1128/jb.178.20.5977-5988.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Prakash S., Prakash L. Nucleotide excision repair in yeast. Mutat. Res. Mol. Mech. Mutagen. 2000;451:13–24. doi: 10.1016/S0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- 146.Coin F., Oksenych V., Egly J.M. Distinct Roles for the XPB/p52 and XPD/p44 Subcomplexes of TFIIH in Damaged DNA Opening during Nucleotide Excision Repair. Mol. Cell. 2007;26:245–256. doi: 10.1016/j.molcel.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 147.Oksenych V., De Jesus B.B., Zhovmer A., Egly J.M., Coin F. Molecular insights into the recruitment of TFIIH to sites of DNA damage. EMBO J. 2009;28:2971–2980. doi: 10.1038/emboj.2009.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Habraken Y., Sung P., Prakash L., Prakash S. Yeast excision repair gene RAD2 encodes a single-stranded DNA endonuclease. Nature. 1993;366:365–368. doi: 10.1038/366365a0. [DOI] [PubMed] [Google Scholar]
- 149.Bardwell A.J., Bardwell L., Tomkinson A.E., Friedberg E.C. Specific Cleavage of Model Recombination and Repair Intermediates by the Yeast Rad1-Rad10 DNA Endonuclease. Science. 1994;265:2082–2085. doi: 10.1126/science.8091230. [DOI] [PubMed] [Google Scholar]
- 150.Ogi T., Limsirichaikul S., Overmeer R.M., Volker M., Takenaka K., Cloney R., Nakazawa Y., Niimi A., Miki Y., Jaspers N.G., et al. Three DNA Polymerases, Recruited by Different Mechanisms, Carry Out NER Repair Synthesis in Human Cells. Mol. Cell. 2010;37:714–727. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 151.Wu X., Braithwaite E., Wang Z. DNA ligation during excision repair in yeast cell-free extracts is specifically catalyzed by the CDC9 gene product. Biochemistry. 1999;38:2628–2635. doi: 10.1021/bi982592s. [DOI] [PubMed] [Google Scholar]
- 152.Fitch M.E., Nakajima S., Yasui A., Ford J.M. In Vivo Recruitment of XPC to UV-induced Cyclobutane Pyrimidine Dimers by the DDB2 Gene Product. J. Biol. Chem. 2003;278:46906–46910. doi: 10.1074/jbc.M307254200. [DOI] [PubMed] [Google Scholar]
- 153.Kapetanaki M.G., Guerrero-Santoro J., Bisi D.C., Hsieh C.L., Rapić-Otrin V., Levine A.S. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl. Acad. Sci. USA. 2006;103:2588–2593. doi: 10.1073/pnas.0511160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Guerrero-Santoro J., Kapetanaki M.G., Hsieh C.L., Gorbachinsky I., Levine A.S., Rapić-Otrin V. The cullin 4B-Based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res. 2008;68:5014–5022. doi: 10.1158/0008-5472.CAN-07-6162. [DOI] [PubMed] [Google Scholar]
- 155.Wang H., Zhai L., Xu J., Joo H.Y., Jackson S., Erdjument-Bromage H., Tempst P., Xiong Y., Zhang Y. Histone H3 and H4 Ubiquitylation by the CUL4-DDB-ROC1 Ubiquitin Ligase Facilitates Cellular Response to DNA Damage. Mol. Cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 156.Scrima A., Koníčková R., Czyzewski B.K., Kawasaki Y., Jeffrey P.D., Groisman R., Nakatani Y., Iwai S., Pavletich N.P., Thomä N.H. Structural Basis of UV DNA-Damage Recognition by the DDB1-DDB2 Complex. Cell. 2008;135:1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.De Weerd-Kastelein E.A., Keijzer W., Bootsma D. Genetic heterogeneity of xeroderma pigmentosum demonstrated by somatic cell hybridization. Nat. New Biol. 1972;238:80–83. doi: 10.1038/newbio238080a0. [DOI] [PubMed] [Google Scholar]
- 158.Bailly V., Sommer C.H., Sung P., Prakash L., Prakash S. Specific complex formation between proteins encoded by the yeast DNA repair and recombination genes RAD1 and RAD10. Proc. Natl. Acad. Sci. USA. 1992;89:8273–8277. doi: 10.1073/pnas.89.17.8273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Haber J.E. Mating-Type Genes and MAT Switching in Saccharomyces cerevisiae. Genetics. 2012;191:33–64. doi: 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nasmyth K. The determination of mother cell-specific mating type of switching in yeast by a specific regulator of HO transcription. EMBO J. 1987;6:243. doi: 10.1002/j.1460-2075.1987.tb04745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jensen R.E., Herskowitz I. Directionality and regulation of cassette substitution in yeast. Cold Spring Harb. Symp. Quant. Biol. 1984;49:97–104. doi: 10.1101/SQB.1984.049.01.013. [DOI] [PubMed] [Google Scholar]
- 162.Herskowitz I., Jensen R.E. Putting the HO gene to work: Practical uses for mating-type switching. Methods Enzymol. 1991;194:132–146. doi: 10.1016/0076-6879(91)94011-Z. [DOI] [PubMed] [Google Scholar]
- 163.White C.I., Haber J.E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Raveh D., Hughes S.H., Shafer B.K., Strathern J.N. Analysis of the HO-cleaved MAT DNA intermediate generated during the mating type switch in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 1989;220:33–42. [PubMed] [Google Scholar]
- 165.Hicks W.M., Yamaguchi M., Haber J.E. Real-time analysis of double-strand DNA break repair by homologous recombination. Proc. Natl. Acad. Sci. USA. 2011;108:3108–3115. doi: 10.1073/pnas.1019660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Aylon Y., Liefshitz B., Bitan-Banin G., Kupiec M. Molecular Dissection of Mitotic Recombination in the Yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;23:1403–1417. doi: 10.1128/MCB.23.4.1403-1417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Holmes A.M., Haber J.E. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 1999;96:415–424. doi: 10.1016/S0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- 168.Ira G., Malkova A., Liberi G., Foiani M., Haber J.E. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/S0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lisby M., Rothstein R., Mortensen U.H. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lisby M., Mortensen U.H., Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat. Cell Biol. 2003;5:572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- 171.Wolner B., Van Komen S., Sung P., Peterson C.L. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell. 2003;12:221–232. doi: 10.1016/S1097-2765(03)00242-9. [DOI] [PubMed] [Google Scholar]
- 172.Sugawara N., Wang X., Haber J.E. In Vivo Roles of Rad52, Rad54, and Rad55 Proteins in Rad51-Mediated Recombination. Mol. Cell. 2003;12:209–219. doi: 10.1016/S1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 173.Resnick M.A., Martin P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. MGG Mol. Gen. Genet. 1976;143:119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- 174.Pâques F., Haber J.E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/MMBR.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mimitou E.P., Symington L.S. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhu Z., Chung W.H., Shim E.Y., Lee S.E., Ira G. Sgs1 Helicase and Two Nucleases Dna2 and Exo1 Resect DNA Double-Strand Break Ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Beernink H.T., Morrical S.W. RMPs: Recombination/replication mediator proteins. Trends Biochem. Sci. 1999;24:385–389. doi: 10.1016/S0968-0004(99)01451-6. [DOI] [PubMed] [Google Scholar]
- 178.Shinohara A., Shinohara M., Ohta T., Matsuda S., Ogawa T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells. 1998;3:145–156. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- 179.New J.H., Sugiyama T., Zaitseva E., Kowalczykowski S.C. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 180.Gasior S.L., Wong A.K., Kora Y., Shinohara A., Bishop D.K. Rad52 associates with RPA and functions with Rad55 and Rad57 to assemble meiotic recombination complexes. Genes Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Heyer W.D., Li X., Rolfsmeier M., Zhang X.P. Rad54: The Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34:4115. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Solinger J.A., Heyer W.D. Rad54 protein stimulates the postsynaptic phase of Rad51 protein-mediated DNA strand exchange. Proc. Natl. Acad. Sci. USA. 2001;98:8447–8453. doi: 10.1073/pnas.121009898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.McVey M., Khodaverdian V.Y., Meyer D., Cerqueira P.G., Heyer W.-D. Eukaryotic DNA Polymerases in Homologous Recombination. Annu. Rev. Genet. 2016;50:393–421. doi: 10.1146/annurev-genet-120215-035243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sugiyama T., Kowalczykowski S.C. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- 185.Sugiyama T., Kantake N., Wu Y., Kowalczykowski S.C. Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. EMBO J. 2006;25:5539. doi: 10.1038/sj.emboj.7601412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sugiyama T., New J.H., Kowalczykowski S.C. DNA annealing by Rad52 Protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl. Acad. Sci. USA. 1998;95:6049. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gangloff S., Mcdonald J.P., Bendixen C., Arthur L., Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol. Cell. Biol. 1994;14:8391–8398. doi: 10.1128/MCB.14.12.8391-8398.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bennett R.J., Sharp J.A., Wang J.C. Purification and Characterization of the Sgs1 DNA Helicase Activity of Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:9644–9650. doi: 10.1074/jbc.273.16.9644. [DOI] [PubMed] [Google Scholar]
- 189.Bennett R.J., Keck J.L., Wang J.C. Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S. cerevisiae. J. Mol. Biol. 1999;289:235–248. doi: 10.1006/jmbi.1999.2739. [DOI] [PubMed] [Google Scholar]
- 190.Walker J.R., Corpina R.A., Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 191.Clerici M., Mantiero D., Guerini I., Lucchini G., Longhese M.P. The Yku70–Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep. 2008;9:810. doi: 10.1038/embor.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mimitou E.P., Symington L.S. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29:3358. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hopfner K.P., Craig L., Moncalian G., Zinkel R.A., Usui T., Owen B.A.L., Karcher A., Henderson B., Bodmer J.L., McMurray C.T., et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 194.Daley J.M., Vander Laan R.L., Suresh A., Wilson T.E. DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J. Biol. Chem. 2005;280:29030–29037. doi: 10.1074/jbc.M505277200. [DOI] [PubMed] [Google Scholar]
- 195.Pardo B., Ma E., Marcand S. Mismatch Tolerance by DNA Polymerase Pol4 in the Course of Nonhomologous End Joining in Saccharomyces cerevisiae. Genetics. 2006;172:2689. doi: 10.1534/genetics.105.053512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wilson T.E., Grawunder U., Lieber M.R. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 197.Herrmann G., Lindahl T., Schär P. Saccharomyces cerevisiae LIF1: A function involved in DNA double-strand break repair related to mammalian XRCC4. EMBO J. 1998;17:4188. doi: 10.1093/emboj/17.14.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chen X., Tomkinson A.E. Yeast Nej1 Is a Key Participant in the Initial End Binding and Final Ligation Steps of Nonhomologous End Joining. J. Biol. Chem. 2011;286:4931. doi: 10.1074/jbc.M110.195024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Mourrain L., Boissonneault G. Dna repair in haploid context. Int. J. Mol. Sci. 2021;22:12418. doi: 10.3390/ijms222212418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ma Y., Pannicke U., Schwarz K., Lieber M.R. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/S0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 201.Mao Z., Bozzella M., Seluanov A., Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 2008;7:2902–2906. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mathiasen D.P., Lisby M. Cell cycle regulation of homologous recombination in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014;38:172–184. doi: 10.1111/1574-6976.12066. [DOI] [PubMed] [Google Scholar]
- 203.Symington L.S., Gautier J. Double-Strand Break End Resection and Repair Pathway Choice. Annu. Rev. Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 204.Kramer K.M., Brock J.A., Bloom K., Moore J.K., Haber J.E. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol. Cell. Biol. 1994;14:1293. doi: 10.1128/MCB.14.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ooi S.L., Shoemaker D.D., Boeke J.D. A DNA Microarray-Based Genetic Screen for Nonhomologous End-Joining Mutants in Saccharomyces cerevisiae. Science. 2001;294:2552–2556. doi: 10.1126/science.1065672. [DOI] [PubMed] [Google Scholar]
- 206.Game J.C., Mortimer R.K. A genetic study of X-ray sensitive mutants in yeast. Mutat. Res. Mol. Mech. Mutagen. 1974;24:281–292. doi: 10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- 207.Al-Khodairy F., Fotou E., Sheldrick K.S., Griffiths D.J.F., Lehmann A.R., Carr A.M. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol. Biol. Cell. 1994;5:147. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Walworth N., Davey S., Beach D. Fission yeast chkl protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 209.Bennett C.B., Lewis L.K., Karthikeyan G., Lobachev K.S., Jin Y.H., Sterling J.F., Snipe J.R., Resnick M.A. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 2001;29:426–434. doi: 10.1038/ng778. [DOI] [PubMed] [Google Scholar]
- 210.Hanway D., Chin J.K., Xia G., Oshiro G., Winzeler E.A., Romesberg F.E. Previously uncharacterized genes in the UV- and MMS-induced DNA damage response in yeast. Proc. Natl. Acad. Sci. USA. 2002;99:10605–10610. doi: 10.1073/pnas.152264899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Clikeman J.A., Khalsa G.J., Barton S.L., Nickoloff J.A. Homologous recombinational repair of double-strand breaks in yeast is enhanced by MAT heterozygosity through yKU-dependent and -independent mechanisms. Genetics. 2001;157:579–589. doi: 10.1093/genetics/157.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Åström S.U., Okamura S.M., Rine J. Yeast cell-type regulation of DNA repair. Nature. 1999;397:310. doi: 10.1038/16833. [DOI] [PubMed] [Google Scholar]
- 213.Lee S.E., Pâques F., Sylvan J., Haber J.E. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol. 1999;9:767–770. doi: 10.1016/S0960-9822(99)80339-X. [DOI] [PubMed] [Google Scholar]
- 214.Mercier G., Berthault N., Touleimat N., Képès F., Fourel G., Gilson E., Dutreix M. A haploid-specific transcriptional response to irradiation in Saccharomyces cerevisiae. Nucleic Acids Res. 2005;33:6635–6643. doi: 10.1093/nar/gki959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Shrivastav M., De Haro L.P., Nickoloff J.A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 216.Simpson-Lavy K.J., Bronstein A., Kupiec M., Johnston M. Cross-talk between carbon metabolism and the DNA damage response in S. cerevisiae. Cell Rep. 2015;12:1865. doi: 10.1016/j.celrep.2015.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zhou C., Elia A.E.H., Naylor M.L., Dephoure N., Ballif B.A., Goel G., Xu Q., Ng A., Chou D.M., Xavier R.J., et al. Profiling DNA damage-induced phosphorylation in budding yeast reveals diverse signaling networks. Proc. Natl. Acad. Sci. USA. 2016;113:E3667–E3675. doi: 10.1073/pnas.1602827113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Yi C., Tong J., Lu P., Wang Y., Zhang J., Sun C., Yuan K., Xue R., Zou B., Li N., et al. Formation of a Snf1-Mec1-Atg1 Module on Mitochondria Governs Energy Deprivation-Induced Autophagy by Regulating Mitochondrial Respiration. Dev. Cell. 2017;41:59–71.e4. doi: 10.1016/j.devcel.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 219.Corcoles-Saez I., Dong K., Johnson A.L., Waskiewicz E., Costanzo M., Boone C., Cha R.S. Essential Function of Mec1, the Budding Yeast ATM/ATR Checkpoint-Response Kinase, in Protein Homeostasis. Dev. Cell. 2018;46:495–503.e2. doi: 10.1016/j.devcel.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 220.Jagadeesan S.K., Potter T., Al-gafari M., Hooshyar M., Hewapathirana C.M., Takallou S., Hajikarimlou M., Burnside D., Samanfar B., Moteshareie H., et al. Discovery and identification of genes involved in DNA damage repair in yeast. Gene. 2022;831:146549. doi: 10.1016/j.gene.2022.146549. [DOI] [PubMed] [Google Scholar]
- 221.Sanford E.J., Comstock W.J., Faça V.M., Vega S.C., Gnügge R., Symington L.S., Smolka M.B. Phosphoproteomics reveals a distinctive Mec1/ATR signaling response upon DNA end hyper-resection. EMBO J. 2021;40:e104566. doi: 10.15252/embj.2020104566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Lao J.P., Ulrich K.M., Johnson J.R., Newton B.W., Vashisht A.A., Wohlschlegel J.A., Krogan N.J., Toczyski D.P. The Yeast DNA Damage Checkpoint Kinase Rad53 Targets the Exoribonuclease, Xrn1. G3 Genes Genomes Genet. 2018;8:3931–3944. doi: 10.1534/g3.118.200767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Mojumdar A., Adam N., Cobb J.A. Nej1 interacts with Sae2 at DNA double-stranded breaks to inhibit DNA resection. J. Biol. Chem. 2022;298:101937. doi: 10.1016/j.jbc.2022.101937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kissling V.M., Reginato G., Bianco E., Kasaciunaite K., Tilma J., Cereghetti G., Schindler N., Lee S.S., Guérois R., Luke B., et al. Mre11-Rad50 oligomerization promotes DNA double-strand break repair. Nat. Commun. 2022;13:2374. doi: 10.1038/s41467-022-29841-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hamza A., Tammpere E., Kofoed M., Keong C., Chiang J., Giaever G., Nislow C., Hieter P. Complementation of yeast genes with human genes as an experimental platform for functional testing of human genetic variants. Genetics. 2015;201:1263–1274. doi: 10.1534/genetics.115.181099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Aldred P.M.R., Borts R.H. Humanizing mismatch repair in yeast: Towards effective identification of hereditary non-polyposis colorectal cancer alleles. Biochem. Soc. Trans. 2007;35:1525–1528. doi: 10.1042/BST0351525. [DOI] [PubMed] [Google Scholar]
- 227.Barbari S.R., Kane D.P., Moore E.A., Shcherbakova P.V. Functional analysis of cancer-associated DNA polymerase ε variants in Saccharomyces cerevisiae. G3 Genes Genomes Genet. 2018;8:1019–1029. doi: 10.1534/G3.118.200042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kim C., Yang J., Jeong S.H., Kim H., Park G.H., Shin H.B., Ro M.J., Kim K.Y., Park Y.J., Kim K.P., et al. Yeast-based assays for characterization of the functional effects of single nucleotide polymorphisms in human DNA repair genes. PLoS ONE. 2018;13:e0193823. doi: 10.1371/journal.pone.0193823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Spugnesi L., Balia C., Collavoli A., Falaschi E., Quercioli V., Caligo M.A., Galli A. Effect of the expression of BRCA2 on spontaneous homologous recombination and DNA damage-induced nuclear foci in Saccharomyces cerevisiae. Mutagenesis. 2013;28:187–195. doi: 10.1093/mutage/ges069. [DOI] [PubMed] [Google Scholar]
- 230.Duan M., Selvam K., Wyrick J.J., Mao P. Genome-wide role of Rad26 in promoting transcription-coupled nucleotide excision repair in yeast chromatin. Proc. Natl. Acad. Sci. USA. 2020;117:18608–18616. doi: 10.1073/pnas.2003868117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Peritore M., Reusswig K.U., Bantele S.C.S., Straub T., Pfander B. Strand-specific ChIP-seq at DNA breaks distinguishes ssDNA versus dsDNA binding and refutes single-stranded nucleosomes. Mol. Cell. 2021;81:1841–1853.e4. doi: 10.1016/j.molcel.2021.02.005. [DOI] [PubMed] [Google Scholar]
- 232.Gnügge R., Symington L.S. Efficient DNA double-strand break formation at single or multiple defined sites in the Saccharomyces cerevisiae genome. Nucleic Acids Res. 2020;48:e115. doi: 10.1093/nar/gkaa833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Vyas V.K., Bushkin G.G., Bernstein D.A., Getz M.A., Sewastianik M., Barrasa M.I., Bartel D.P., Fink G.R. New CRISPR Mutagenesis Strategies Reveal Variation in Repair Mechanisms among Fungi. mSphere. 2018;3:e00154-18. doi: 10.1128/mSphere.00154-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Lemos B.R., Kaplan A.C., Bae J.E., Ferrazzoli A.E., Kuo J., Anand R.P., Waterman D.P., Haber J.E. CRISPR/Cas9 cleavages in budding yeast reveal templated insertions and strand-specific insertion/deletion profiles. Proc. Natl. Acad. Sci. USA. 2018;115:E2010–E2047. doi: 10.1073/pnas.1716855115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Resnicks M.A. Genetic control of radiation sensitivity in Saccharomyces cerevisiae. Genetics. 1969;62:519–531. doi: 10.1093/genetics/62.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Snow R. Mutants of Yeast Sensitive to Ultraviolet Light. J. Bacteriol. 1967;94:571. doi: 10.1128/jb.94.3.571-575.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Schultzhaus Z., Chen A., Shuryak I., Wang Z. The Transcriptomic and Phenotypic Response of the Melanized Yeast Exophiala dermatitidis to Ionizing Particle Exposure. Front. Microbiol. 2021;11:609996. doi: 10.3389/fmicb.2020.609996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.