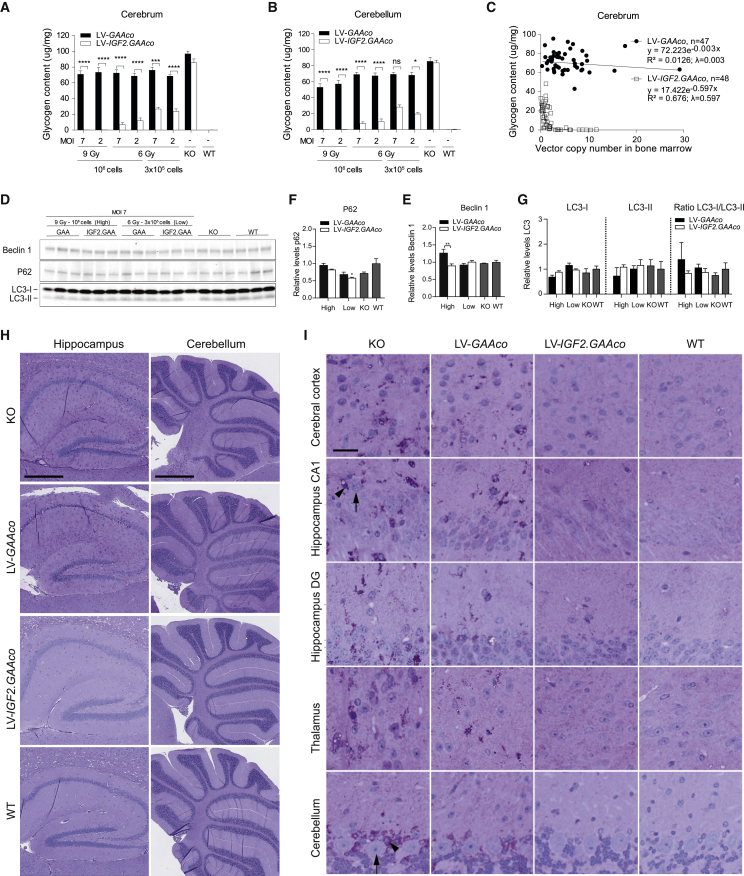
Figure 5.
Gene therapy with LV-IGF2.GAAco results in correction in brain
(A and B) Glycogen content in control and gene-therapy-treated mice in cerebrum and cerebellum. MOI, multiplicity of infection; Gy, gray. (C) Correlation between VCN and glycogen clearance in cerebrum. λ, exponential decay constant. VCN is not normalized for chimerism. (D–G) Immunoblot analysis for beclin 1, p62, and LC3 in cerebrum after gene therapy. Age-matched WT and Gaa−/− animals were taken as control. Density levels of beclin 1 (E), p62 (F), or LC3 (G) were quantified from (D) (total protein control is shown in Figure S5C). (E–G) Quantification density values are relative to WT level (set as 1). (H and I) PAS staining analysis in cerebrum and cerebellum after high-dose gene therapy. An overview of the stained area is presented in (H). Scale bar, 0.5 mm (hippocampus) or 1 mm (cerebellum). Representative images at indicated areas are shown in (I). Scale bar, 50 μm. Black arrows point to pyramidal neurons (in hippocampus CA1) and Purkinje cells (in cerebellum). Black arrowheads refer to the glial cells. DG, dentate gyrus; KO, untreated Gaa−/− mice; WT, untreated WT mice. Data are presented as means ± SEM. (A and B) Data are analyzed by two-way ANOVA with Bonferroni’s correction, using vector (LV-GAAco or LV-IGF2.GAAco) and gene therapy dose as nominal predictor variables. Significant results are indicated by brackets. Results of multiple comparison analysis are reported in Table S3. Western blot quantifications (E–G) are analyzed by one-way ANOVA followed by Bonferroni’s multiple testing. Significance versus WT is shown; other significant comparisons are indicated by brackets. LV-GAAco and LV-IGF2.GAAco, n = 7; KO, n = 5; and WT, n = 5. (E–G) n = 3. (H and I) LV-GAAco and LV-IGF2.GAAco, n = 3 per group; KO, n = 2; and WT, n = 2. ns, not significant; ∗p ≤ 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p ≤ 0 .0001.