FIGURE 1.
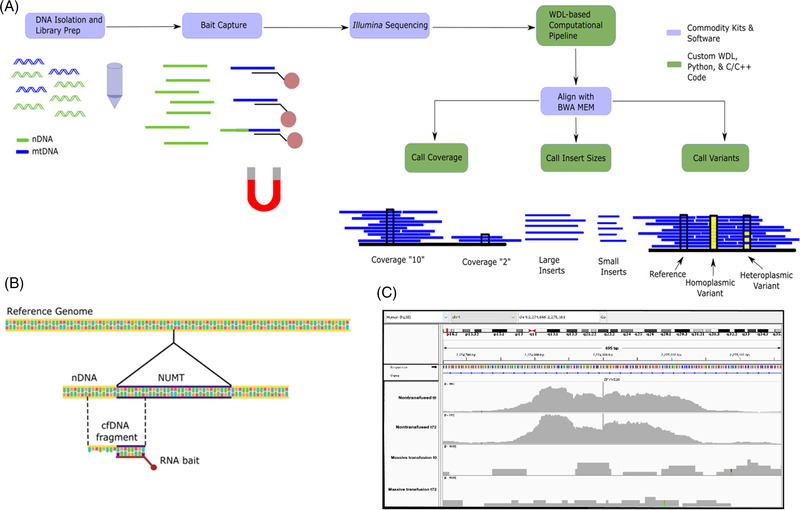
RNA target bait‐capture and bioinformatics protocol and nuclear mitochondrial (NUMT) identification. (A) DNA is isolated from plasma or tissue. In the figure, nuclear DNA (nDNA) and mitochondrial DNA (mtDNA) are denoted by colour. DNA isolation and library preparation are applied to all sample DNA, regardless of nuclear or mitochondrial origin. A target bait‐capture kit consists of biotinylated RNA probes complementary to the mitochondrial genome. The probes efficiently bind mtDNA but can also bind homologous NUMT, as illustrated by the DNA fragment half coloured as mitochondrial and half coloured as nuclear. Once enriched, samples are pooled and sequenced on a standard Illumina instrument. From there, a Workflow Description Language pipeline aligns the reads to the whole genome –nuclear and mitochondrial. Three custom C/C++ programs built on the htslib library then call mitochondrial and nuclear coverage, insert (the size of a fragment after end repair and sequencing adapter ligation), and variant calling. (B) Schematic depiction of how target‐bait capture also leads to the sequencing of flanking regions of polymorphic NUMTs. (C) Integrated Genome Viewer (IGV) histograms depicting a specific polymorphic NUMT in a nontransfused patient whereas the second patient lacks this insertion at either t0 or t72 post‐admittance. Subfigures (A) and (B) were prepared in Inkscape. (C) Prepared from IGV.
