Figure 3. Inactive p53 dimers are held in place by residues in the DBD.
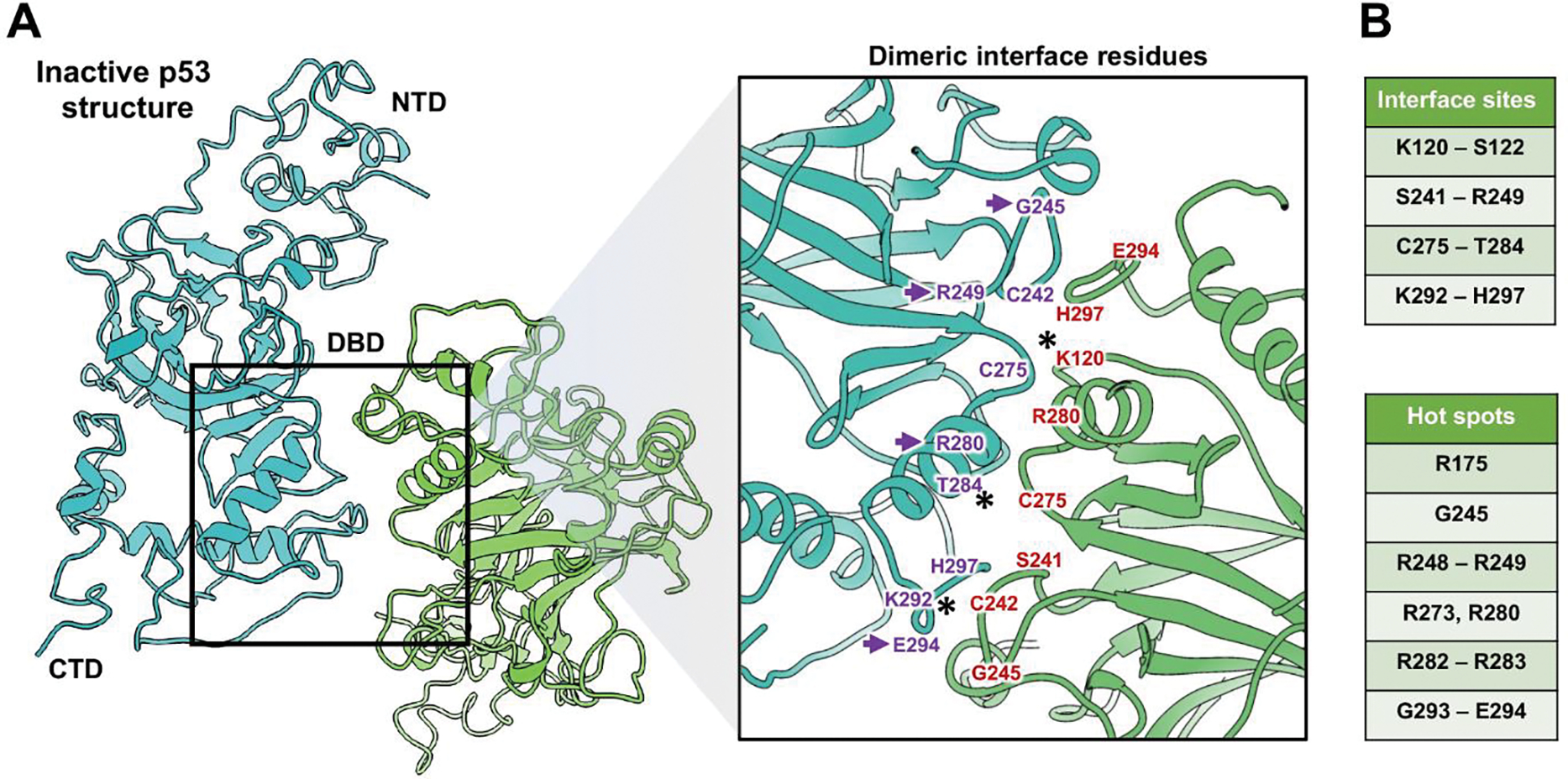
(A) The p53 dimer interface revealed key residues in the DBD domain that contributed to its structural stability. The NTD, DBD, and CTD are shown for dimer constituents that form an overall “crossed” architecture. A magnified view of the dimer interface shows the spatial relationship between residues. (B) Amino acids in the interface site (purple and red) are listed along with additional residues that are considered mutational hot spots of cancer-relevance. Purple arrows point to hot spot mutations. (*) indicates residues that can be modified by ubiquitination, acetylation (K120 and K292) or by phosphorylation (T284). These modifications may help to modulate the dimer activation state.
