Figure 1.
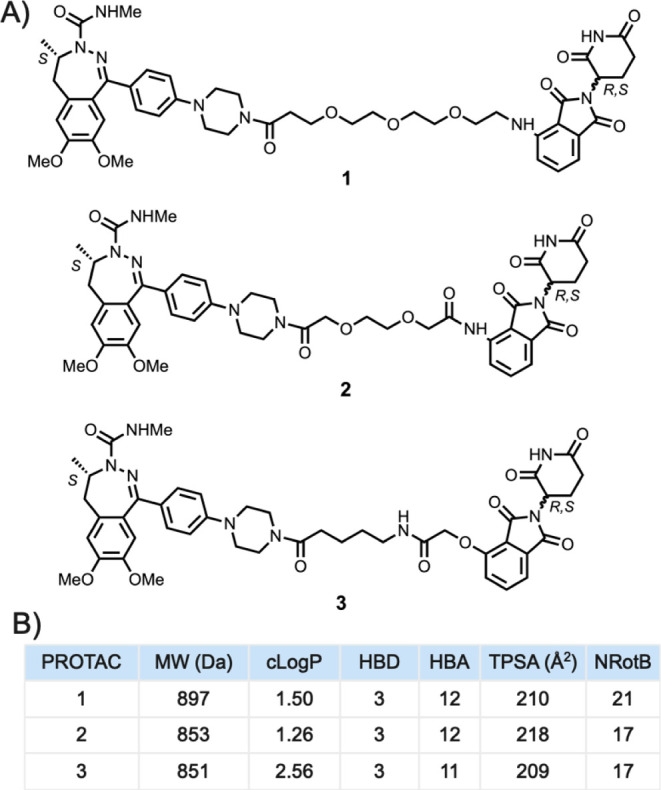
(A) Structures of PROTACs 1–3. The three PROTACs have the same BRD4 and E3 ligase (CRBN) ligands but differ in the structures of their linkers. (B) Descriptors of Lipinski’s rule of 527 and Veber’s rule28 were calculated for PROTACs 1–3 using MOE (version 2019.01). MW, molecular weight; cLog P, calculated lipophilicity; HBD, hydrogen bond donor; HBA, hydrogen bond acceptor; TPSA, topological polar surface area; and NRotB, number of rotatable bonds.
