Fig. 3.
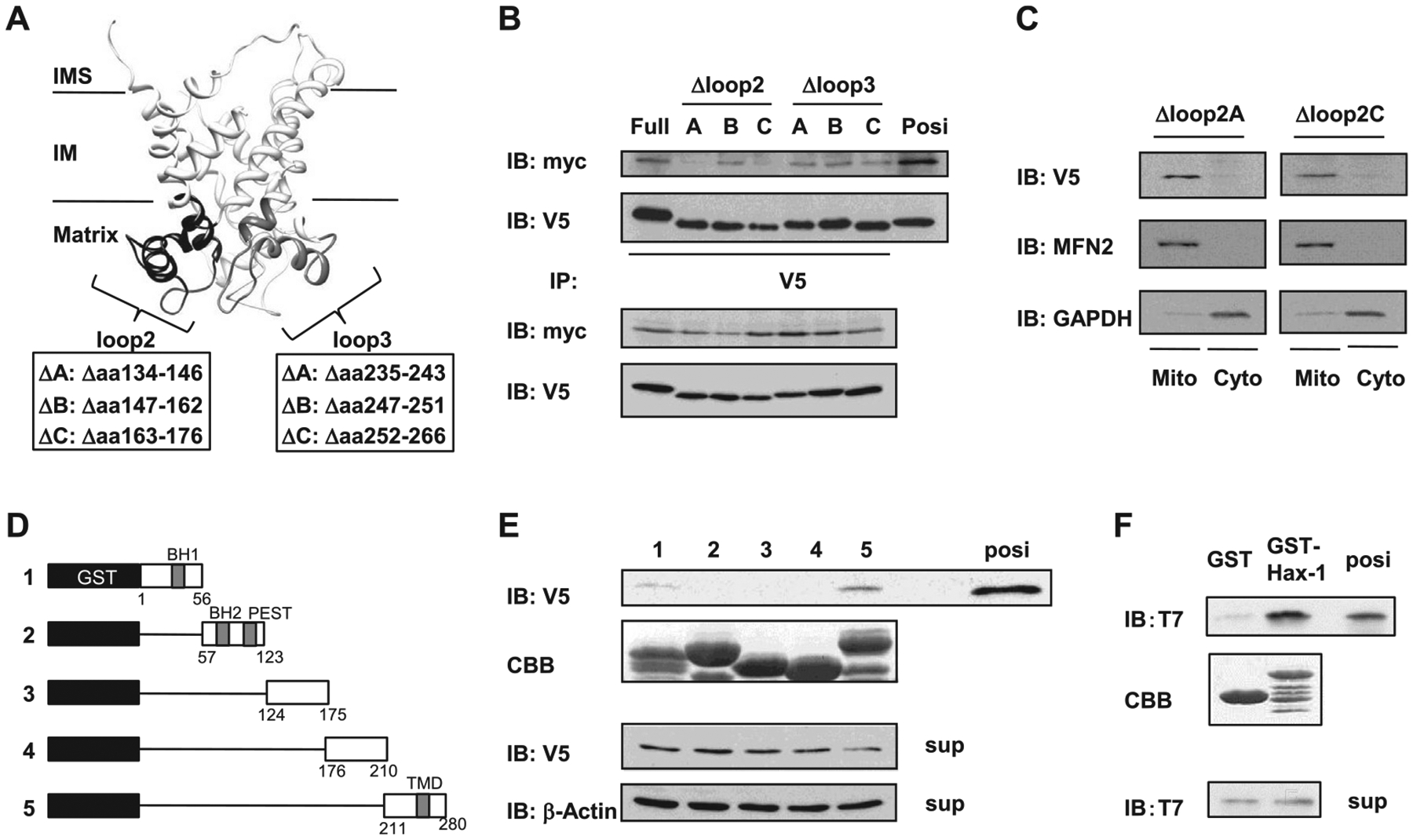
Mutational mapping of the UCP3-Hax-1 binding domain. (A) Speculative structure of mouse UCP3 was rendered with UCSF chimera (17). The UCP3 loop2 domain truncation mutants of UCP3 (UCP3 Δloop2A134–146, Δloop2B147–162, and Δloop2C163–176), and the UCP3 loop3 domain truncation mutants of UCP3 (UCP3 Δloop3A235–243, Δloop3B247–251, and Δloop3C252–266) were constructed as negative controls to determine the binding domain to Hax-1. (B) The loop 2 hydrophilic domain of UCP3 required for binding to Hax-1 was defined using mutagenesis and immunoprecipitation. Cell extracts (100 μg) were immunoprecipitated with anti-V5 antibodies and analyzed by immunoblotting with anti-myc antibodies. (C) Mitochondria and cytosolic extracts (20 μg/lane) were isolated from HEK293 cells transfected with UCP3 mutations and were subjected to SDS-PAGE followed by immunoblotting with anti-V5, anti-mitofusin 2 (MFN2), and anti-GAPDH antibodies. (D) Diagram for expression constructs of (1) GST-Hax-1 residues 1–56, (2) GST-Hax-1 residues 57–123, (3) GST-Hax-1 residues 124–175), (4) GST-Hax-1 residues 176–210), and (5) GST-Hax-1 residues 211–280. (E) The residues of Hax-1 required for direct binding to UCP3 were determined with GST pull-down assays. GST-fusion proteins were purified and stained by Coomassie Brilliant Blue (CBB) as indicated. In HEK293 cell extracts expressing UCP3–V5, the ability of the truncated Hax-1 fusion proteins to bind to UCP3 was analyzed by immunoblotting against V5. (F) Direct protein interactions were determined by incubating purified recombinant T7-tagged UCP3 IML2 domain with GST-Hax-1 or GST. The samples from the GST pull-down assays were subjected to SDS-PAGE and immunoblotting against T7. Purified protein of the UCP3 IML2 domain was used as a positive control.
