Figure 1.
Sequencing and data analysis workflow
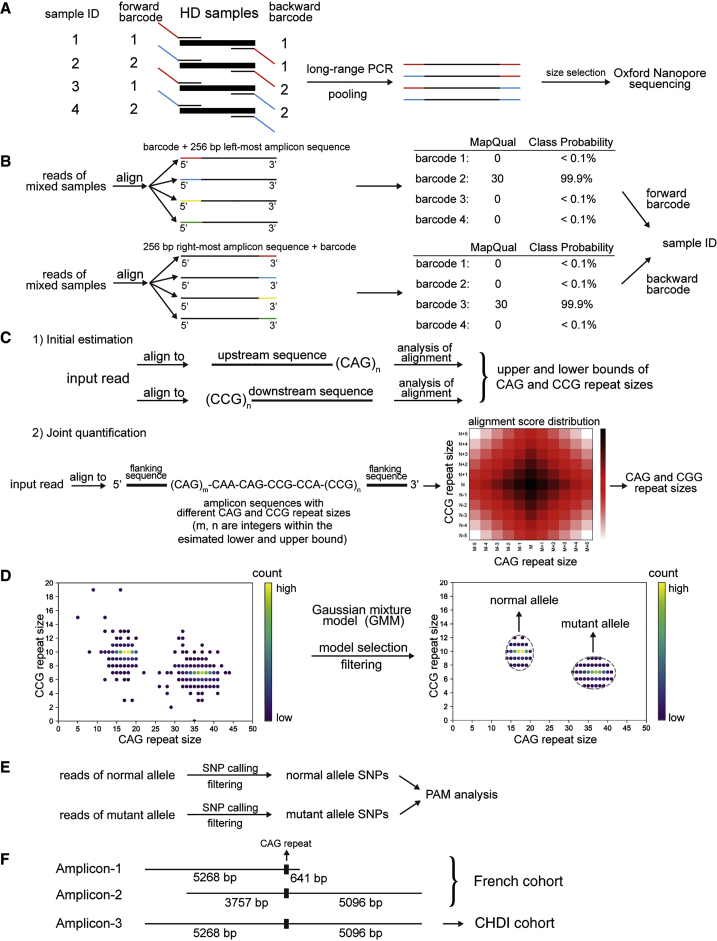
(A) Barcodes were added to both forward and backward PCR primers and the pooled PCR products of multiple samples were sequenced with Oxford nanopore sequencing.
(B) De-multiplexing strategy of NanoBinner. NanoBinner aligns a read to barcode sequences with the 256-bp amplicon sequence immediately next to it. The 256-bp amplicon sequence acts as an anchor so that the matching of barcodes is at the correct position. NanoBinner assigns a read to a barcode if the Phred scale mapping quality score is ≥30.
(C) Joint quantification of CAG and CCG repeat sizes using NanoRepeat. NanoRepeat first estimates the upper and lower bounds of the CAG and CCG repeat sizes separately, and then performs a joint quantification to refine the repeat sizes. In the joint quantification step, NanoRepeat generates a series of template sequences with m CAG repeats and n CCG repeats, where m and n are all integers within the upper and lower bounds determined in the first step. A read was aligned to this series of template sequences. The CAG and CCG repeat sizes that maximize the alignment score was the final estimates of the repeat sizes of the read.
(D) NanoRepeat separates reads using GMM. CAG and CCG repeat sizes are used as input features. The scatterplot shows the CAG and CCG repeat sizes of a typical example. The color of the points indicates the number of reads. Model selection is performed to select the best number of Gaussian models. After filtering outliers, the two alleles are well separated (right). The dashed gray circles are equi-probability surfaces of the fitted Gaussian models where the probability outside the surface is less than 5%.
(E) SNPs detection was performed using longshot. Low quality SNP calls were removed. The effects of on PAMs were examined.
(F) Locations of the PCR amplicons of each cohort. The lengths are the distance to the first nucleotide of the CAG repeat (based on GRCh38).