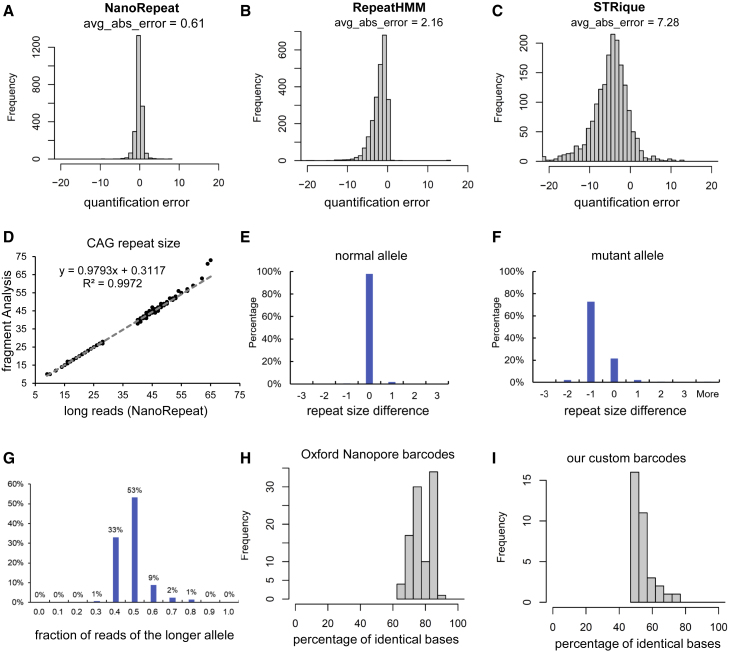
Figure 2.
Assessment of computational methods
(A–C) Benchmarking repeat quantification on 2370 STR regions in the CHM13 genome. The average absolute error (avg_abs_error) of each method is shown.
(D) Scatterplot showing the CAG repeat size quantified by NanoRepeat and PCR-based fragment analysis.
(E and F) Repeat size difference between NanoRepeat’s results and the PCR-based fragment analysis.
(G) Distribution of the fraction of reads of the longer allele.
(H and I) Histogram showing the percentage of identical bases between the barcode sequence and the best aligned sequence in the human reference genome GRCh38. Sequences of 96 Oxford Nanopore barcodes were obtained from the online documentation of the PCR Barcoding Expansion Pack.