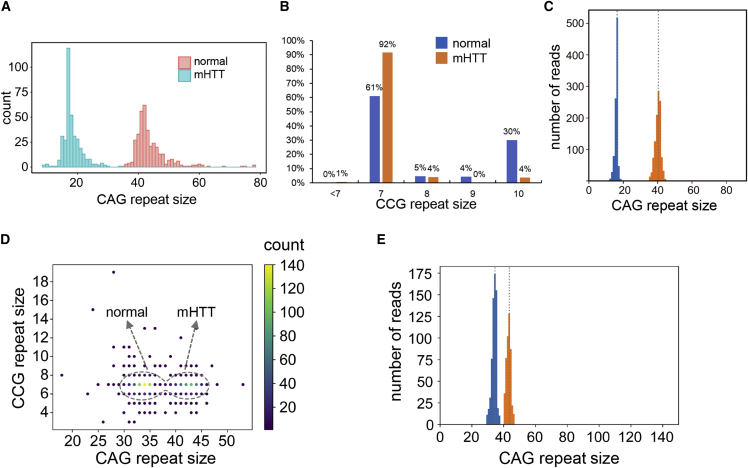
Figure 3.
CAG and CCG repeats detected from HD samples
(A) Distribution of CAG repeat size of the French cohort.
(B) The normal and mHTT allele showed distinct distribution in CCG repeat sizes.
(C) CAG repeat size distribution of a typical HD sample. The reads derived from the normal allele (blue) and the mHTT allele (orange) are well separated. The estimated repeat sizes are marked by vertical gray lines.
(D) Scatterplot of a sample with a disease-causing allele (CAG repeat size = 43) and an intermediate allele (CAG repeat size = 34). The dashed gray circle is an equi-probability surface of the fitted Gaussian models where the probability outside the surface is less than 5%.
(E) Distribution of CAG repeat size of the same sample shown in (D). Reads that are not confidently classified are removed. The two alleles are well separated (blue, normal allele; orange, mHTT allele). The estimated repeat sizes are marked by vertical gray lines.