Figure 5.
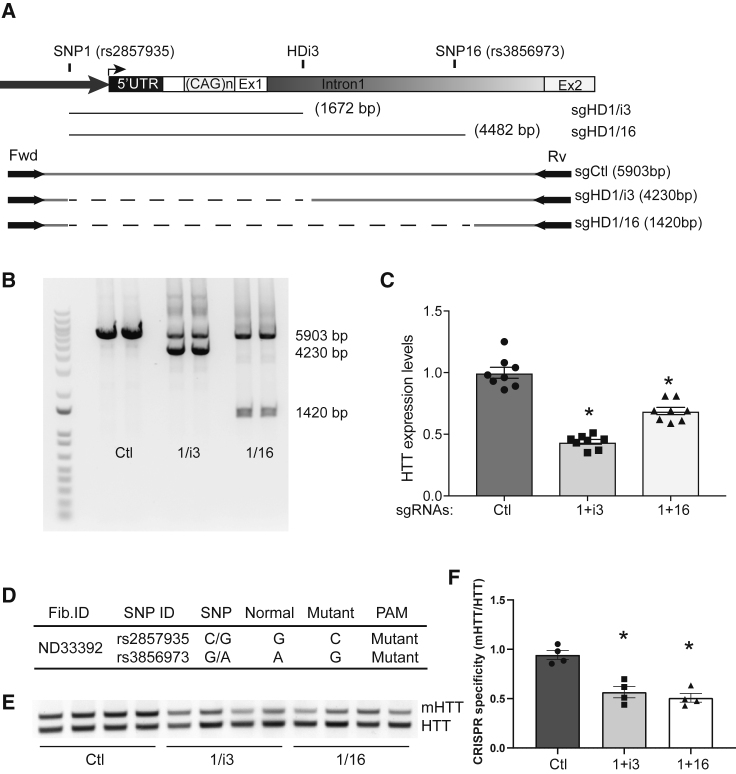
Allele-specific cleavage mediated by SNP1 and SNP16
(A) Cartoon depicting the relative position of SNP1- and SNP16-dependent PAMs flanking HTT exon-1 and one common PAM within HTT intron-1 (HDi3). PCR primer positions and estimated sizes of the targeted deleted sequences are indicated. 1, i3, and 16 are sgRNAs targeting PAMs at SNP1, HDi3, and SNP16, respectively.
(B) A genomic PCR showing HTT exon-1-targeted deletion mediated by SNP1- and SNP16-dependent PAMs.
(C) qRT-PCR analysis of HTT mRNA levels in HEK293 cells transfected with CRISPR enzymes targeting SNP1 and SNP16 (n = 8).
(E) The haplotype of the ND33392 fibroblast cell line and the corresponding PAMs.
(F) A semiquantitative PCR reaction showing the reduction of the mHTT allele in a fibroblast cell line (ND33392) transfected with SaCas9 targeting SNP1 and SNP16.
(C) and (F) The results are mean ± SEM relative to the control group. ∗p < 0.05, one-way ANOVA followed by Bonferroni’s post-hoc.