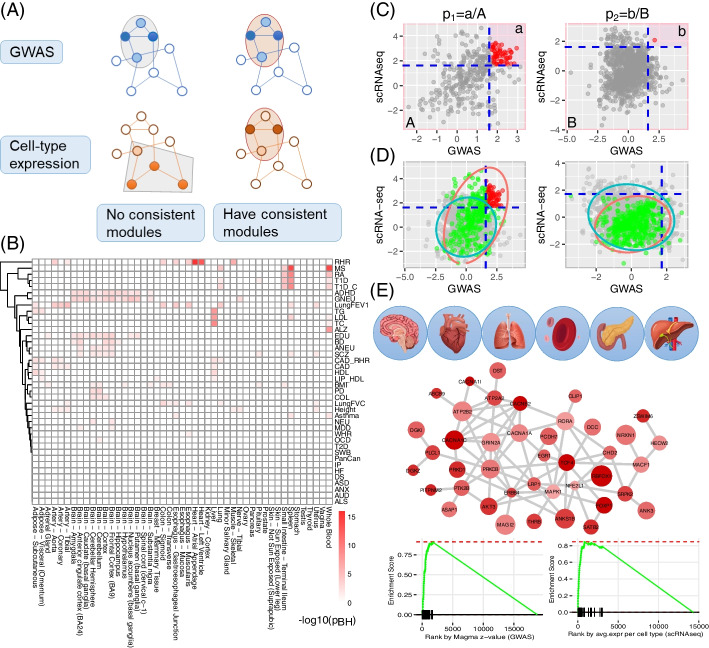
Fig. 1.
Analysis framework to decode trait-associated tissues and cell types. A Illustration of GWAS and cell-type expression integration at the cellular level. B Tissue-specific enrichment analysis of the traits. The color reflects the significance level [− log10(pBH)]. C Demonstration of the proportional test. D Demonstration of a case showing the association between GWAS and cell type transcriptome (left) and another case without such an association (right). In each figure, a dot indicates a module, with its GWAS-based score shown on the x-axis and its scRNA-seq score shown on the y-axis. The gray dots are random modules from the randomization process. The green and red dots are modules from the real data whereas the red ones indicate significance. Cyan and red circles indicate the 95% confidence interval (CI) of the random modules and the actual modules, respectively. The horizontal and vertical dash lines indicate nominal significance (z = 1.96). E Illustration of disease subnetworks and the enrichment result of the component genes in each of the two heterogeneous data sets