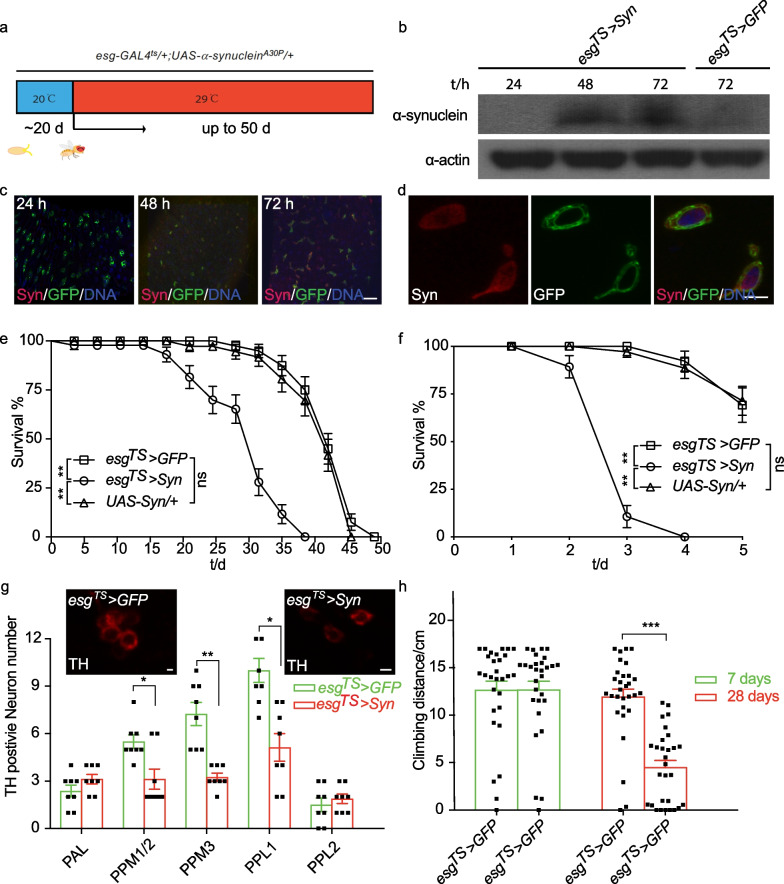
Fig. 1.
Intestinal overexpression of α-syn recapitulates Parkinson’s disease pathology in Drosophila. a Diagram of the temporal expression of the α-syn transgene in the Drosophila intestine. Temperature-controlled expression of UAS-Syn or UAS-GFP was achieved by co-expression of esg-GAL4 with a temperature-sensitive version of the GAL4 inhibitor Gal80TS, in which the expression of UAS-synuclein was inactivated and induced at 18 °C and 29 °C, respectively. We refer to the two genotypes as esgTS > Syn (w; esg-Gal4, UAS-GFP/+; UAS-Syn/tubulin-Gal80TS) and esgTS > GFP (w; esg-Gal4, UAS-GFP/+; UAS-GFP/tubulin-Gal80TS). b Western blot showing the expression of human α-syn in the midgut of adult flies after 48 h at 29 °C. α-Actin was used as a loading control. c Immunostaining showing the expression of human α-syn in fly intestinal stem cells (Scale bar, 1 mm). α-Syn expression in intestinal stem cells increased following a shift to 29 °C. d Representative images showing exogenous α-syn in both cytoplasm and nucleus (Scale bar, 1.5 μm). Red: α-syn; green: GFP; blue: DNA. e Decreased lifespan of intestinal PD flies. Adult flies were cultured as described in panel a and the percentage of living male flies was calculated to monitor lifespan. f Survival rates of intestinal PD flies challenged with rotenone. Sucrose solution containing 7.5 mM rotenone was used for survival assay. g Intestinal overexpression of α-syn causes an age-dependent dopaminergic (DA) neuron loss. Numbers of TH-positive DA neurons in five clusters of each genotype were counted and analyzed (bottom). Representative images of TH-positive DA neurons in the PPM1 cluster of 28-day-old control or intestinal α-syn-expressing flies (top). Scale bars, 0.2 μm. h Expression of α-syn in the midgut leads to progressive locomotor deficits. For the locomotion assay, climbing distances of each genotype of males at different time points are plotted as bar graphs. Mean ± SEM; P-values of survival curves were calculated using log-rank tests (using total fly numbers), and for category graphs using one-way ANOVA with a Bonferroni multiple-comparison test. *P < 0.05; **P < 0.01; ***P < 0.001