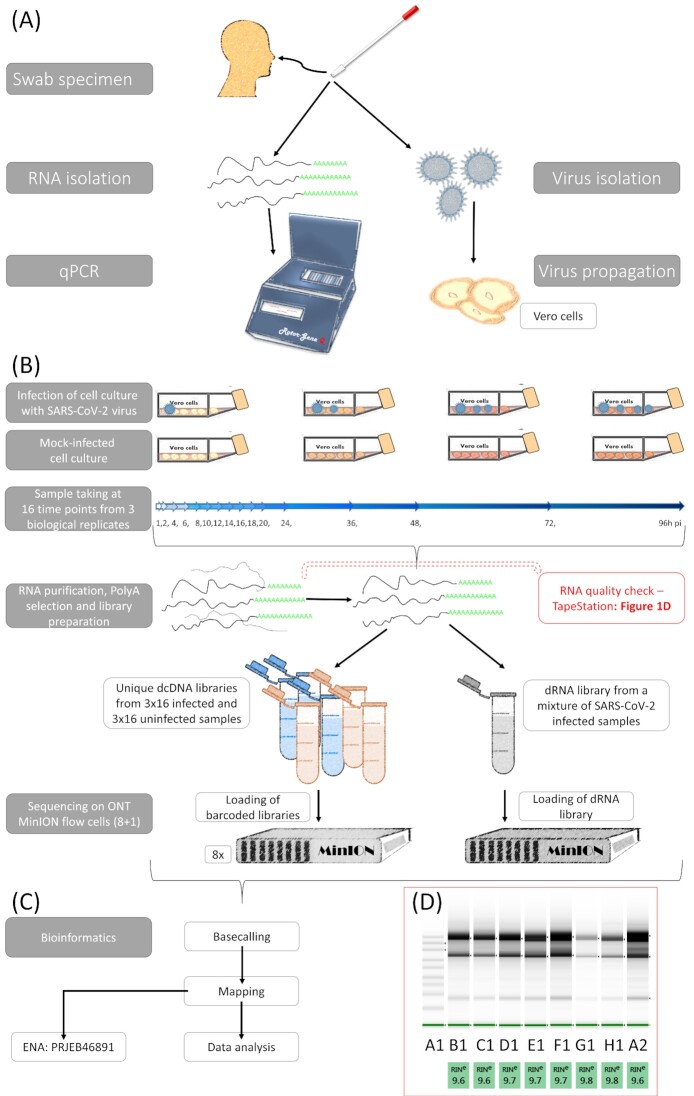
Figure 1:
Schematic representation of the workflow applied in this project. (A) Isolation and detection of a Hungarian isolate of the SARS-CoV-2 virus. The sample was collected from a human nasopharyngeal swab. The SARS-CoV-2 infection was validated by reverse transcription PCR using the RNA extracted from the sample. The virus was isolated from the sample and was maintained on Vero cells. (B) Experimental workflow of the study. Vero cells were infected with SARS-CoV-2 and the cells were incubated at 37°C for 1, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 24, 36, 48, 72, and 96 hours post infection. Uninfected control cells were also propagated. Each time-point experiment was carried out in 3 biological replicates. RNAs were purified from the samples, which was followed by the preparation of libraries and then sequencing using direct cDNA and direct RNA methods. Altogether, 9 MinION flow cells (ONT) were used for this study. (C) Bioinformatics workflow. The ONT's Guppy basecaller was used to identify the base sequence of the obtained reads, and then they were aligned to the viral and host reference genomes by using the minimap2 mapper. Statistical data were generated with seqtools [25] and a custom R-workflow [33]. (D) Quality of RNA samples was detected with a TapeStation 2200 system with RNA ScreenTape. TapeStation gel image shows that intact, high-quality RNAs were isolated from the samples and used for sequencing. The image shows the following samples: A1: marker; B1: 8-hour postinfection (pi) sample C; 12-hour pi sample A; 16-hour pi sample A; 18-hour pi sample B, 20-hour pi sample C; 36-hour pi sample A; 48-hour pi sample A; 96-hour pi sample B.