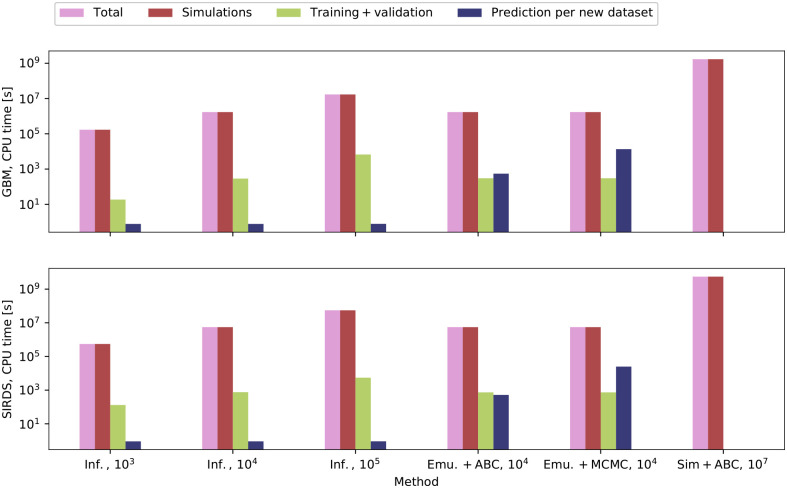
Fig 6. Required CPU time across different methods for our cancer CA model (upper panel) and the epidemiological SIRDS model (lower panel).
The light magenta bars show the total time required for each inference technique. The remaining three bars specify the time required for the simulations involved (red), the time required for training and validation (yellow), and the time that is needed to infer the parameters for a single set of observations (blue). The number of required simulations is specified in the labels. We include the NN direct inference machine based on three different sizes of the training set. Moreover, we include the NN emulator in tandem with the rejection ABC and MCMC algorithms. The ABC accepts the best 104 among 107 randomly drawn samples. The ensemble MCMC uses 8 walkers, drawing 20,000 samples for each walker. For the MCMC, we average over the CPU times for cases a and b. For comparison, we include the estimated time required for running the ABC with 107 simulations. Note that the ordinate is logarithmic. Note also that the time needed to run any given simulation depends on the number of agents and the model parameters.