FIGURE 1.
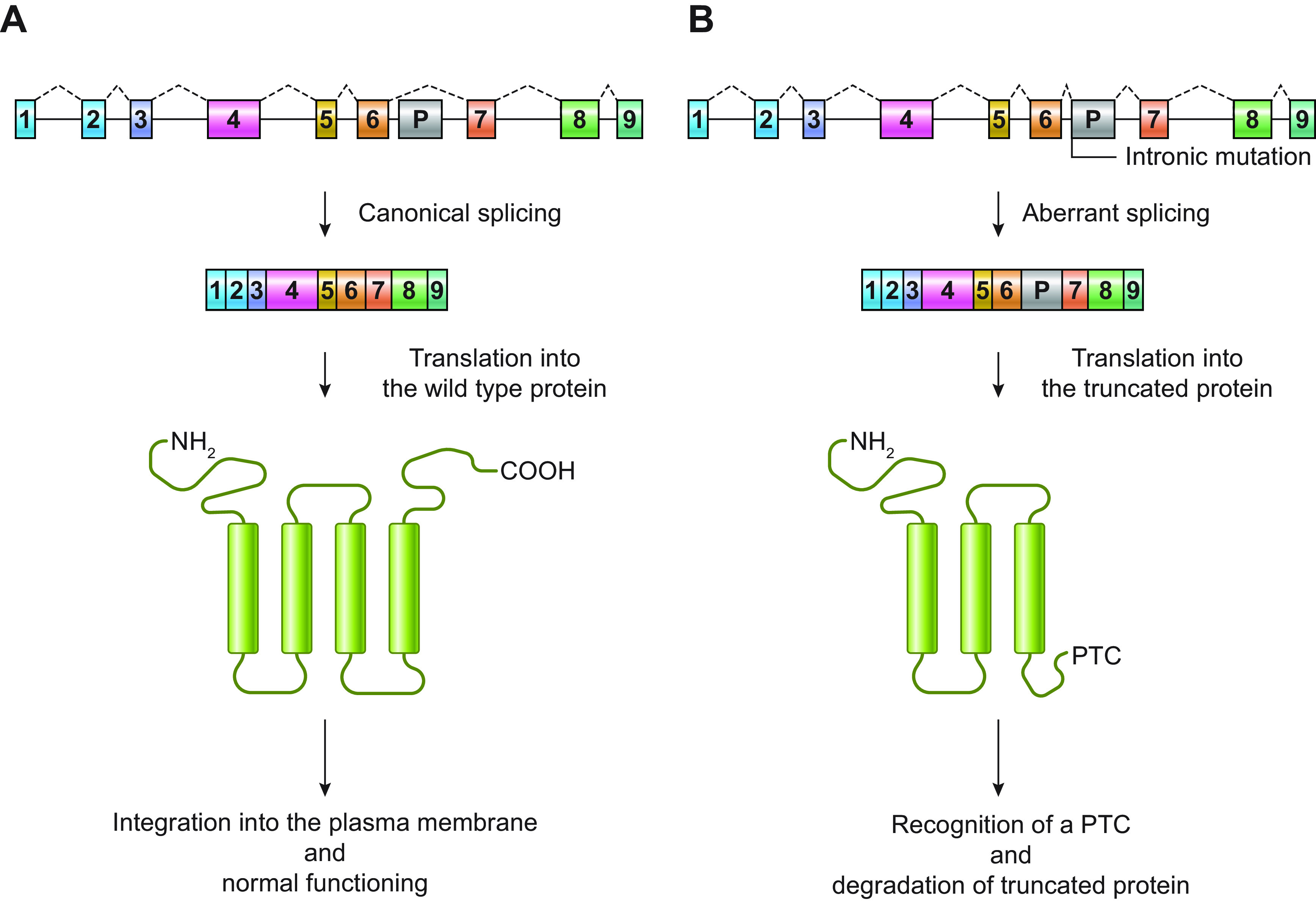
Schematic representation of an example of “poison exon”-mediated protein degradation. A shows a hypothetical gene encoding a transmembrane protein with 4 transmembrane segments. The gene comprises 9 coding exons (1–9) and a potential poison exon (P). In the canonical splicing, the poison exon is not included in the mRNA, which is translated into the wild-type protein. After translation, the protein is correctly integrated into the plasma membrane, where it exerts its normal function. In B, the presence of an intronic mutation, which can introduce a novel splicing acceptor site, activates an exonic splicing enhancer (ESE; i.e., a sequence that promotes the inclusion of an exon in an mRNA) or disrupts an exonic splicing silencer (ESS; i.e., a sequence that inhibits the inclusion of an exon in an mRNA), promoting the inclusion of the poison exon in the mRNA. The poison exon alters protein amino acid sequence and introduces a premature stop codon (PTC). The PTC is recognized by cellular surveillance systems, and the mutant protein is degraded.
