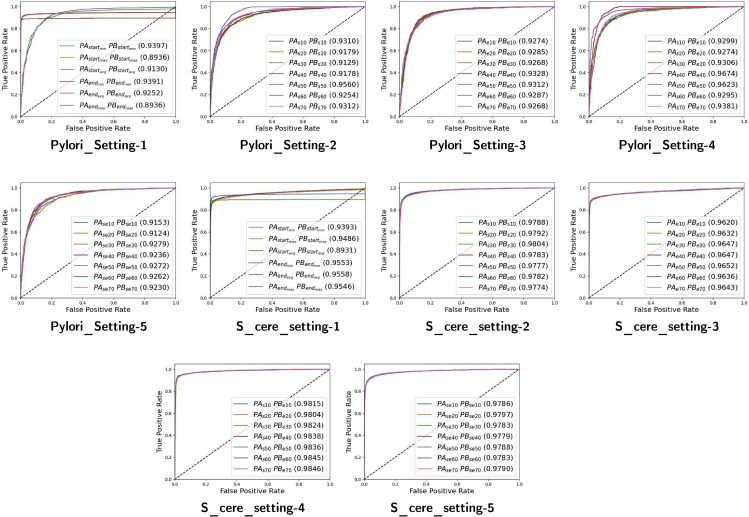
Figure 3.
Impact of 5 different settings on the performance of proposed ADH-PPI approach across 2 different S.cerevisiae and H.pyloir datasets for the task of PPI prediction in terms of area under receiver operating characteristics
Setting 1 is based on traditional copy padding, sequence truncation, and hybrid approaches. Setting 2, 3, 4, and 5 are based on subsequences criteria where x number of amino acids from staring and ending regions of Protein A and Protein B are taken. The value of x varies from 10 to 70 amino acids with the difference of 10 amino acids. Setting 2 takes x number of amino acids from starting region of Protein A and ending region of Protein B. Setting 3 takes x number of amino acids from ending region of Protein A and Protein B. Setting 4 takes x number of amino acids from starting region of Protein A and Protein B. Setting 5 takes x number of amino acids from starting and ending region of Protein A and starting and ending region of Protein B.