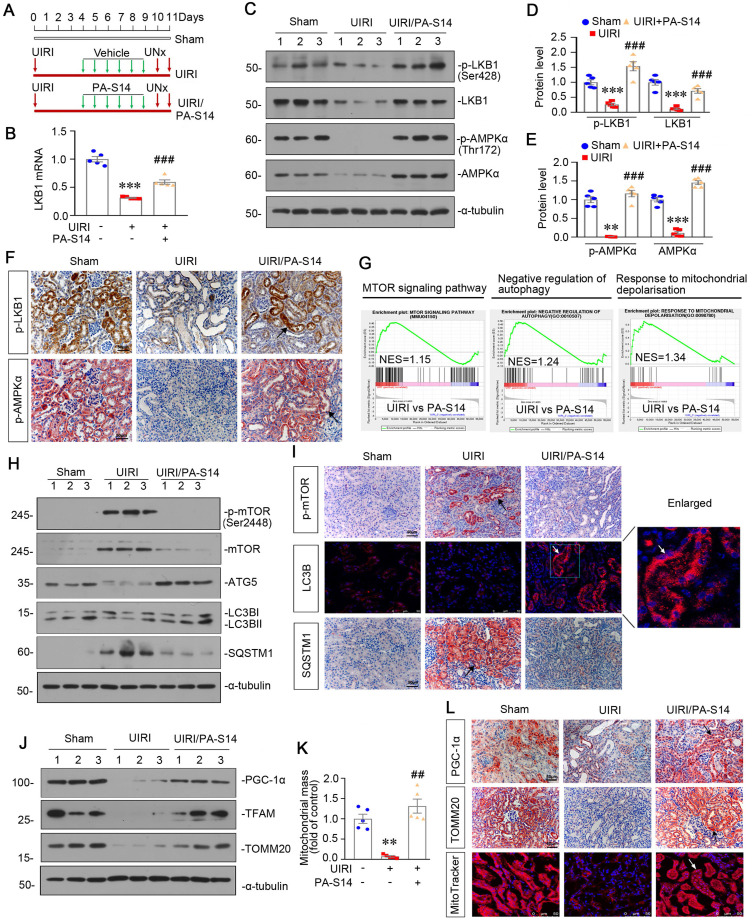
Figure 5.
PA-S14 promotes autophagy and mitochondrial biogenesis in UIRI mice via activation of LKB1-AMPK signaling. (A) Diagram shows the experimental design. Red arrows indicate the time points undergoing UIRI, UNx (uni-nephrectomy) and sacrifice, respectively. Green arrows indicate PA-S14 treatment (1.0 mg/kg body weight). (B) Quantitative PCR result showing renal expression of LKB1 in different groups as indicated. ***P < 0.001 versus sham controls; ###P < 0.001 versus UIRI (n = 5). (C-E) Representative western blot and quantitative data showing renal expression of p-LKB1, LKB1, p-AMPKα, and AMPKα in different groups. Numbers (1-3) indicate each individual animal in a given group. **P < 0.01, ***P < 0.001 versus sham controls; ###P < 0.001 versus UIRI (n = 5). (F) Representative micrographs show the p-LKB1 and p-AMPKα expression in different groups as indicated. Kidney sections were stained with antibodies against p-LKB1 and p-AMPKα, respectively. Arrows indicate positive staining. Scale bar, 50 µm. (G) GSEA enrichment analysis showing that mTOR signaling pathway, negative regulation of autophagy and response to mitochondrial depolarization were enriched in UIRI. NES, normalized enrichment score; Nominal p-value < 0.05 or FDR q-value < 0.25. (H) Representative western blot showing renal expression of the p-mTOR, mTOR, ATG5, LC3BII/I ratio and SQSTM1 in different groups. Numbers (1-3) indicate each individual animal in a given group. (I) Representative micrographs show renal expression of p-mTOR, LC3B and SQSTM1. Paraffin kidney sections were stained with antibodies against p-mTOR and SQSTM1. The frozen kidney sections were stained with an antibody against LC3B. Arrows indicate positive staining. Scale bar, 50 µm. (J) Western blotting analyses show renal expression of PGC‐1α, TFAM and TOMM20 in different groups. Numbers (1-3) represent different individual animals in a given group. (K) Quantification of renal mitochondrial mass is shown. Mitochondrial mass was determined by the fluorescence intensity of MitoTracker deep red staining normalized to DAPI. **P < 0.01 versus sham controls; ##P < 0.01 versus UIRI (n = 5). (L) Representative micrographs show the expression of PGC-1α, TOMM20 and mitochondrial mass. Paraffin kidney sections were stained with antibodies against PGC-1α and TOMM20. The frozen kidney sections were stained with MitoTracker deep red (3 µm) probe. DAPI (4′,6‐diamidino‐2‐phenylindole) was used to stain the nuclei (blue). Arrows indicate positive staining. Scale bar, 50 µm.