Use of rapid tests for diagnosis of COVID-19 is now commonplace, but questions remain regarding their performance characteristics compared with those of polymerase chain reaction testing. The role of sequential rapid testing in improving sensitivity is of great interest.
Visual Abstract. Rapid Antigen Test Performance for Delta vs. Omicron SARS-CoV-2 Variants.
Use of rapid tests for diagnosis of COVID-19 is now commonplace, but questions remain regarding their performance characteristics compared with those of polymerase chain reaction testing. The role of sequential rapid testing in improving sensitivity is of great interest.
Abstract
Background:
It is important to document the performance of rapid antigen tests (Ag-RDTs) in detecting SARS-CoV-2 variants.
Objective:
To compare the performance of Ag-RDTs in detecting the Delta (B.1.617.2) and Omicron (B.1.1.529) variants of SARS-CoV-2.
Design:
Secondary analysis of a prospective cohort study that enrolled participants between 18 October 2021 and 24 January 2022. Participants did Ag-RDTs and collected samples for reverse transcriptase polymerase chain reaction (RT-PCR) testing every 48 hours for 15 days.
Setting:
The parent study enrolled participants throughout the mainland United States through a digital platform. All participants self-collected anterior nasal swabs for rapid antigen testing and RT-PCR testing. All Ag-RDTs were completed at home, whereas nasal swabs for RT-PCR were shipped to a central laboratory.
Participants:
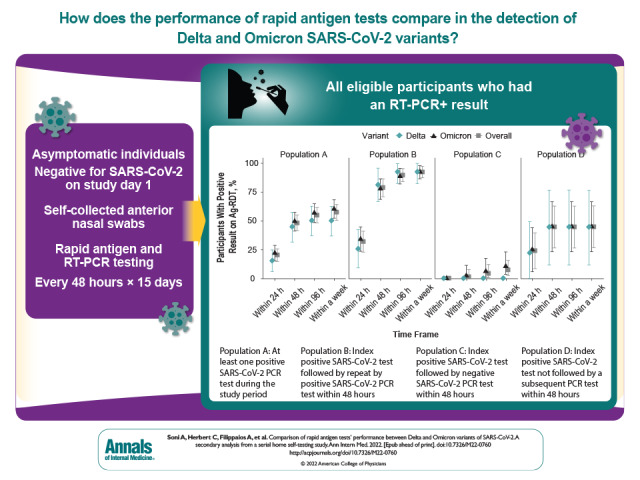
Of 7349 participants enrolled in the parent study, 5779 asymptomatic persons who tested negative for SARS-CoV-2 on day 1 of the study were eligible for this substudy.
Measurements:
Sensitivity of Ag-RDTs on the same day as the first positive (index) RT-PCR result and 48 hours after the first positive RT-PCR result.
Results:
A total of 207 participants were positive on RT-PCR (58 Delta, 149 Omicron). Differences in sensitivity between variants were not statistically significant (same day: Delta, 15.5% [95% CI, 6.2% to 24.8%] vs. Omicron, 22.1% [CI, 15.5% to 28.8%]; at 48 hours: Delta, 44.8% [CI, 32.0% to 57.6%] vs. Omicron, 49.7% [CI, 41.6% to 57.6%]). Among 109 participants who had RT-PCR–positive results for 48 hours, rapid antigen sensitivity did not differ significantly between Delta- and Omicron-infected participants (48-hour sensitivity: Delta, 81.5% [CI, 66.8% to 96.1%] vs. Omicron, 78.0% [CI, 69.1% to 87.0%]). Only 7.2% of the 69 participants with RT-PCR–positive results for shorter than 48 hours tested positive by Ag-RDT within 1 week; those with Delta infections remained consistently negative on Ag-RDTs.
Limitation:
A testing frequency of 48 hours does not allow a finer temporal resolution of the analysis of test performance, and the results of Ag-RDTs are based on self-report.
Conclusion:
The performance of Ag-RDTs in persons infected with the SARS-CoV-2 Omicron variant is not inferior to that in persons with Delta infections. Serial testing improved the sensitivity of Ag-RDTs for both variants. The performance of rapid antigen testing varies on the basis of duration of RT-PCR positivity.
Primary Funding Source:
National Heart, Lung, and Blood Institute of the National Institutes of Health.
Accurate and accessible testing for SARS-CoV-2 is a critical tool for the timely identification of infection to inform isolation recommendations, prevent transmission, and facilitate early initiation of therapy to reduce disease progression (1). Rapid antigen tests (Ag-RDTs) for COVID-19 show great promise as a testing method that is easy to use, accessible, and cost-effective (2). Results from Ag-RDTs are available within minutes of sample collection, compared with hours to days for results from reverse transcriptase polymerase chain reaction (RT-PCR) tests. The U.S. federal government launched a program in January 2022 to distribute a half billion Ag-RDTs at no cost to U.S. residents in an effort to improve the country's ability to respond to a surge in COVID-19 cases (3).
Rapid antigen tests have lower sensitivity than RT-PCR tests for detecting SARS-CoV-2 (4); however, sensitivity can be improved through serial testing (5). Existing data on the performance of Ag-RDTs predate the emergence of the Omicron (B.1.1.529) variant, which has mutations throughout the SARS-CoV-2 genome. In particular, mutations in the nucleocapsid gene may lead to protein conformational changes that affect the target binding site of Ag-RDTs. This could theoretically alter the performance of Ag-RDTs in detecting this variant (6–9). The rapid global emergence and dominance of the Omicron variant highlight the importance of understanding the performance of Ag-RDTs in real-world settings.
The urgent need to reassess the performance of Ag-RDTs in detecting the SARS-CoV-2 Omicron variant is further compounded by early reports that Ag-RDTs have lower sensitivity for the Omicron variant than for other variants (10, 11). Recent reports from analytic studies suggest that Ag-RDT performance does not vary across the Delta (B.1.617.2) and Omicron variants; however, previous studies have not looked at the serial performance of tests or identification of new-onset infections (12–14). This article analyzes Ag-RDT performance for detection of the Delta and Omicron variants of SARS-CoV-2 by comparing the results of Ag-RDTs versus nasal RT-PCR tests when testing participants serially every 48 hours.
Methods
Study Population
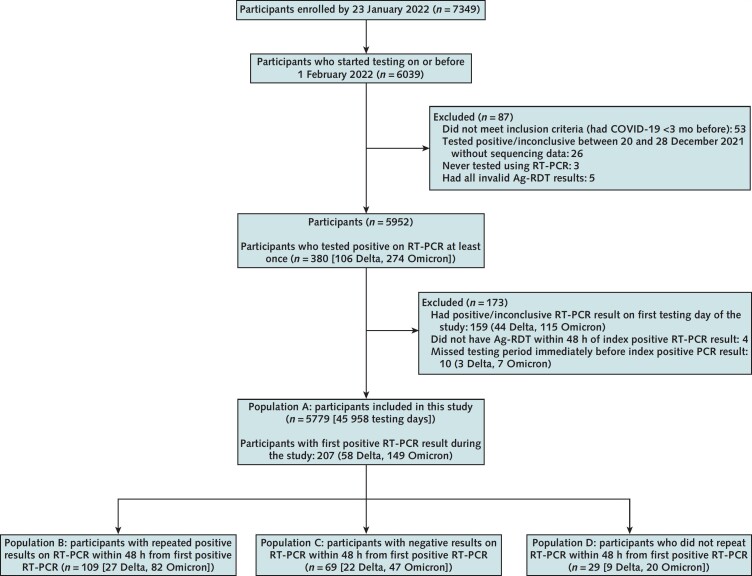
This analysis used data collected in the TUAH (Test Us At Home) study. TUAH is a prospective cohort study that was done by the National Institutes of Health Rapid Acceleration of Diagnostics (RADx) program's Clinical Studies Core; this initiative featured a collaboration among the National Institutes of Health, the U.S. Food and Drug Administration, and University of Massachusetts Chan Medical School. Enrollment occurred from 18 October 2021 to 1 February 2022. Persons older than 2 years residing in any state except Hawaii, Alaska, or Arizona were eligible for TUAH, provided they had access to a smartphone and could receive mail at home. Persons with COVID-19 symptoms in the 14 days before enrollment or a self-reported positive test result for COVID-19 in the previous 3 months were excluded from the study. Study enrollment was self-directed through the study-specific project under the MyDataHelps app (CareEvolution). Participants whose first RT-PCR test in the study had a positive result were excluded from this analysis to allow us to analyze testing performance in the context of RT-PCR positivity onset, as were those who missed a testing period immediately before their index RT-PCR test with positive results (Figure 1). In addition, participants without Ag-RDT results within 48 hours of index RT-PCR positivity were excluded.
Figure 1. Study flow diagram.
5779 participants were included in this study, representing 45 958 participant-days of testing. 207 participants had a new-onset positive result on RT-PCR testing (58 Delta and 149 Omicron) (population A). The subanalysis was based on the result of RT-PCR testing 48 h after the initial positive RT-PCR result (populations B, C, and D). Ag-RDT = rapid antigen test; RT-PCR = reverse transcriptase polymerase chain reaction.
We defined 4 populations in this study. Population A included all eligible participants who had an RT-PCR–positive result. Populations B, C, and D were subsets of population A, defined by the result of the RT-PCR test taken within 48 hours of the index RT-PCR test with positive results, as described in Figure 1: Population B participants had a repeated positive result within 48 hours of the index test, population C had a negative result within 48 hours of the index test, and population D did not have another RT-PCR test within 48 hours of the index test either because of nonadherence or the end of the study period.
The study protocol for the main study was approved by the University of Massachusetts Chan Medical School Institutional Review Board and externally by Western Institutional Review Board. Additional protocol details for TUAH can be found elsewhere (15).
Study Procedures
On enrollment, participants were assigned to 1 of 3 Ag-RDTs with emergency use authorization (BD Veritor At-Home COVID-19 Test, Quidel QuickVue At-Home OTC COVID-19 Test, and Abbott BinaxNOW COVID-19 Antigen Self Test). Participants received the Ag-RDT and the Quest Diagnostics collection kit for COVID-19 by mail at the shipping address provided on enrollment. Participants were asked to self-collect 2 anterior nasal swabs sampled from both nostrils and use 1 swab to complete the Ag-RDT (at home) and 1 for comparator RT-PCR testing (shipped to central laboratory) on the same day roughly every 48 hours for 15 days, as described in Supplement Table 1. Participants were instructed to always collect the Ag-RDT sample first and have at least a 15-minute break before sample collection for the RT-PCR test. Instructions for the tests, specifically for self-collecting and shipping the comparator specimens, were provided as authorized by the Food and Drug Administration. The RT-PCR assay was based on the Roche cobas SARS-CoV-2 assay and had emergency use authorization for use with specimens collected with the Quest Diagnostics collection kit for COVID-19. For participants who tested positive in December or January and had adequate remnant sample, we did whole-genome sequencing of SARS-CoV-2 by amplicon-based next-generation sequencing on extracted RNA. Viral-specific primer sequences and methods of generating the viral genome sequence by consensus were adapted from the ARTIC network.
Variables
The result of an Ag-RDT was based on self-report by the participant in the MyDataHelps app. The RT-PCR result was based on laboratory determination and was considered positive for this analysis if at least 1 of the 2 targets of Roche cobas RT-PCR assays for SARS-CoV-2 was detected. Cycle threshold (Ct) values for the E gene from RT-PCR were used in analyses to quantify viral load. Vaccination history and SARS-CoV-2 infection history were based on self-report using the MyDataHelps app. Case patients were assigned to the Omicron group on the basis of a positive RT-PCR result from a sample collected on 1 January 2022 or later and to the Delta group on the basis of a positive RT-PCR result from a sample collected before 20 December 2021; these cutoff dates were based on sequencing results (Supplement Table 2). Participants who tested positive between 20 and 31 December 2021 were assigned to their respective group on the basis of the sequencing results; those without sequencing results in this period were excluded (Figure 1 and Supplement Table 2).
Statistical Analysis
This is not the prespecified study analysis but was subsequently developed to address an ancillary research question using this unique and comprehensive longitudinal data set (15). Specific analysis related to symptomatic status was not pursued because of overlap with the primary objectives of the parent study (16). Descriptive statistics were calculated at the participant level using tabulation of frequencies for categorical data, and differences were compared using χ2 or Fisher exact tests, depending on the cell sample size. We calculated Ag-RDT sensitivity at different time points for the different populations described in Figure 1. The numerator was based on participants who had at least 1 positive Ag-RDT result in the corresponding time frame since the first positive RT-PCR result (same day, within 48 hours, within 96 hours, or within 1 week). The denominator was based on total number of eligible participants with RT-PCR positivity in each population. We also calculated sensitivity differences for Delta and Omicron. Corresponding 95% CIs for each proportion were calculated using the delta method that uses Taylor linearization. All statistical analyses were done using Stata, version 17.0 (StataCorp).
Role of the Funding Source
This study was funded by the National Institutes of Health RADx Tech program. The funders assisted with study design but had no role in data collection or analysis or the decision to submit the findings for publication.
Results
Cohort Characteristics and RT-PCR Test Results
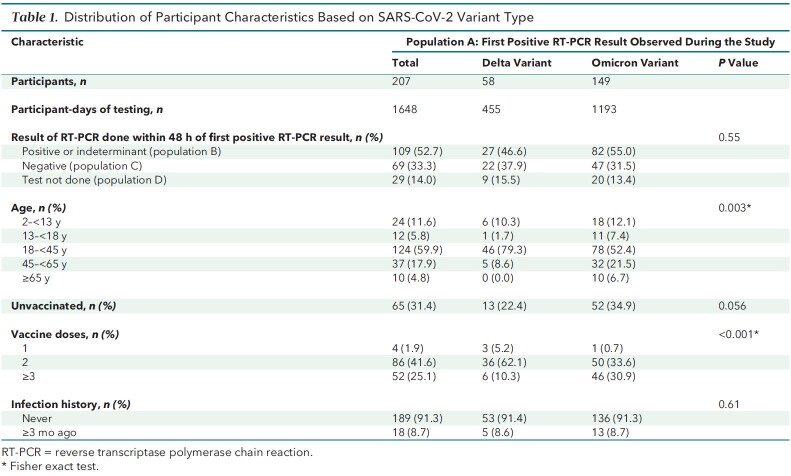
A total of 6039 participants enrolled in the TUAH study and did home-based testing between 21 October 2021 and 1 February 2022. This analysis was limited to 5779 eligible participants (Figure 1). Data from 45 958 participant-days of testing were available from this analytic sample. During the study period, 207 participants (58 Delta, 149 Omicron) had an initial positive result on an RT-PCR test and were classified as population A (Table 1 and Figure 1). Of these participants, 109 (52.6%) had a subsequent positive RT-PCR result within 48 hours of the first positive result (population B), 69 (33.3%) had a subsequent negative RT-PCR result within 48 hours (population C), and 29 (14.0%) did not have an RT-PCR test within 48 hours after their first positive RT-PCR result (population D) (Supplement Table 3). The proportion of persons with singleton positive results on RT-PCR (population C) was similar among participants infected with the Delta (37.9%) and Omicron (31.5%) variants (P = 0.54). Slightly more participants who tested positive on RT-PCR (population A) were unvaccinated during the Omicron period (34.9%) than during the Delta period (22.4%); however, this was not statistically significant (P = 0.056).
Table 1.
Distribution of Participant Characteristics Based on SARS-CoV-2 Variant Type
Time From RT-PCR Positivity to Ag-RDT Positivity Among Delta and Omicron Variants
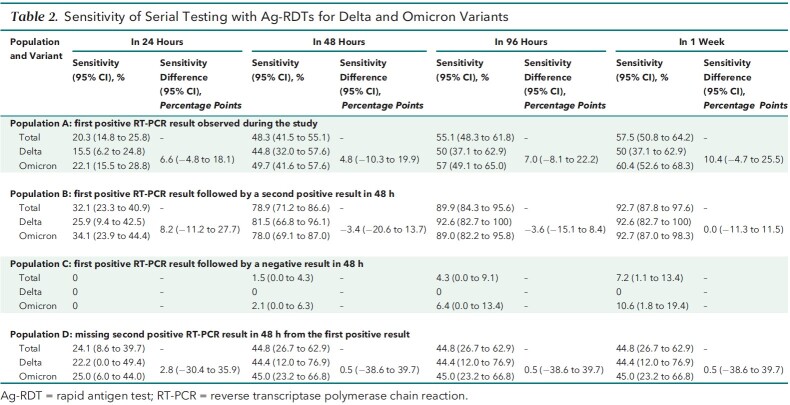
Among the 207 participants in population A whose index positive result on an RT-PCR test was observed during the study period, the sensitivity of Ag-RDTs was 20.3% (95% CI, 14.8% to 25.8%) on the day of the index test and 55.1% (CI, 48.3% to 61.8%) within 48 hours afterward (Table 2 and Figure 2). The proportions of Omicron- and Delta-infected participants who were Ag-RDT–positive on the same day, within 48 hours, within 96 hours, and within a week of the index positive RT-PCR result did not differ significantly.
Table 2.
Sensitivity of Serial Testing with Ag-RDTs for Delta and Omicron Variants
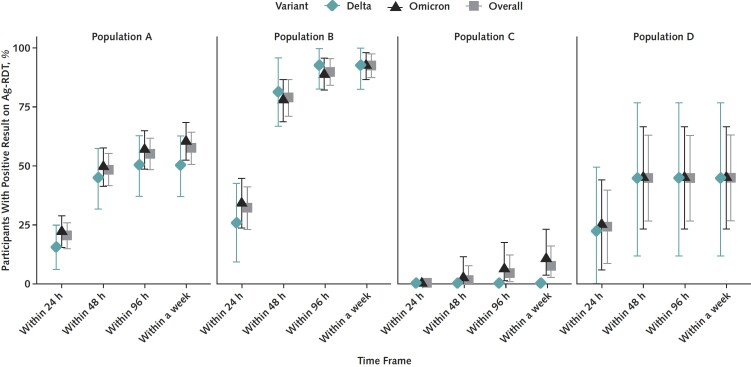
Figure 2. Proportion of participants testing positive by Ag-RDT, by days since initial sample collection for positive RT-PCR result.
Proportion of RT-PCR–positive participants who tested positive by Ag-RDT did not significantly differ between Delta and Omicron infections. Ag-RDT performance improved when tests were used serially. Ag-RDT = rapid antigen test; RT-PCR = reverse transcriptase polymerase chain reaction.
Among participants with at least 2 sequential positive results on RT-PCR tests (population B), 78.9% (CI, 71.2% to 86.6%) and 89.9% (CI, 84.3% to 95.6%) were Ag-RDT–positive within 48 and 96 hours, respectively, from first RT-PCR positivity. Of the 107 participants who were serially positive on RT-PCR for at least 48 hours, similar proportions of Omicron- and Delta-infected participants tested positive on Ag-RDTs within 48 hours from the first positive RT-PCR result (Delta, 81.5% [CI, 66.8% to 96.1%] vs. Omicron, 78.0% [CI, 69.1% to 87.0%]) (Table 2 and Figure 2). The sensitivity of Ag-RDTs among participants with a negative RT-PCR result within 48 hours of the index positive RT-PCR result (that is, population C) was 7.3% (CI, 2.4% to 16.1%) at 1 week. Among the 69 participants in population C, only 5 had a positive Ag-RDT result at some point during the study. Unlike other participants in population C, all 5 of these participants with a positive Ag-RDT result turned serially RT-PCR–positive later in the study period.
Relationship Between Probability of Ag-RDT Positivity and Ct Value Among Delta and Omicron Variants
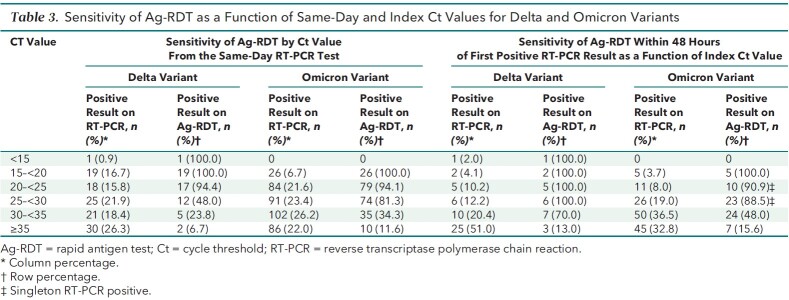
Sensitivity was similar between variants for same-day positivity of Ag-RDTs compared with RT-PCR when the Ct count was less than 30 (Delta, 77.8% vs. Omicron, 89.1%) and for 48-hour positivity of Ag-RDT compared with RT-PCR when the Ct count was less than 30 (Delta, 100% vs. Omicron, 90.5%) (Table 3). Compared with participants infected with the Delta variant, those with Omicron infections had a higher predicted probability of Ag-RDT positivity when the Ct value was lower than 30; however, this difference was not statistically significant.
Table 3.
Sensitivity of Ag-RDT as a Function of Same-Day and Index Ct Values for Delta and Omicron Variants
Discussion
In this analysis of data from 5779 participants that included 45 958 participant-days of Ag-RDT and RT-PCR testing spanning October 2021 to January 2022, we found that Ag-RDT performance for detection of the Omicron variant was not inferior to that of the Delta variant. Of note, overall (in population A), the same-day positivity for Ag-RDTs on onset of RT-PCR positivity was low at 20.3%. Repeated Ag-RDT testing within 48 hours improved this proportion to 48.3% overall. Among the participants who were positive on RT-PCR testing for at least 48 hours, Ag-RDT results were positive for 32.1% on the same day and 78.9% within 48 hours. The phenomenon of singleton RT-PCR positivity (population C) merits further discussion because Ag-RDTs were largely unable to detect the infection, regardless of the variant. Taken together, our findings suggest that Ag-RDTs detect infections similarly for the Delta and Omicron variants, with overall low detection rates on the same day as an initial RT-PCR–positive result and higher detection rates when a second test is used 48 hours after the first.
It is important to consider the following factors when interpreting these results. First, these results present a comparison between Ag-RDT and RT-PCR tests using self-collected nasal swabs. Second, the testing frequency of 48 hours does not allow a finer temporal resolution of the analysis of test performance. Third, the results of Ag-RDTs are based on self-report. However, these limitations are nondifferential and unlikely to bias the comparison of Ag-RDT performance between the Delta and Omicron variants. Furthermore, the data collected from this study illustrate the performance of Ag-RDTs self-collected at home, which more closely resemble the real-world evidence.
Early epidemiologic studies have shown decreased and delayed sensitivity of Ag-RDTs in detecting the Omicron variant in comparison with saliva RT-PCR testing. Adamson and colleagues (10) reported that among 28 people with a positive result on saliva RT-PCR testing with suspected Omicron variant infection and a Ct value lower than 29, none tested positive on nasal Ag-RDT within 24 hours and only 25% tested positive on nasal Ag-RDT within 48 hours. By contrast, among the infections where the Ct value from the initial nasal RT-PCR test with positive results was lower than 30, our study found that the Ag-RDT result was positive within 48 hours in 38 of 42 instances. In the 4 cases in which Ag-RDT positivity was not recorded within 48 hours of the initial RT-PCR positivity with a Ct value lower than 30, all were found to be singleton RT-PCR–positive results, with the subsequent RT-PCR resulting as negative at 48 hours. We also found a sensitivity of 89.1% with Ag-RDT done on the same day as RT-PCR positivity with a Ct count lower than 30 among persons infected with the Omicron variant, which is similar to findings of a separate report that evaluated similar performance among predominantly symptomatic participants (17). The discrepancy between these findings may be explained by the use of saliva RT-PCR instead of nasal RT-PCR as the primary comparator. Marais and colleagues (18) reported that the positive percentage agreement of RT-PCR tests from a nasal swab was higher for the Delta variant (100%) than the Omicron variant (86%); in contrast, that from a saliva sample was higher for the Omicron variant (100%) than the Delta variant (71%). However, considering that saliva PCR tests are not widely available and the typical turnaround time for commercial nasal RT-PCR tests ranges from 36 to 48 hours, our finding suggests that serial use of Ag-RDT may be a viable option for ascertaining SARS-CoV-2 infection status, regardless of Delta or Omicron variant.
The findings from our study reinforce the importance of serial use of Ag-RDTs to overcome the relatively low sensitivity of Ag-RDTs on the first day of RT-PCR positivity. In a previous study of known positives and close contacts, limited sensitivity was observed for an Ag-RDT at a single time point in the early course of infection, but repeated testing every 48 or 72 hours improved sensitivity from lower than 40% to nearly 80% (5). Viral dynamics with Omicron infection may be different, such that there is a more rapid increase in the RNA viral load but a lower peak and shorter clearance phase in comparison with the Delta variant (19). Indeed, we observed a slightly higher proportion of first Ct values less than 30 for Omicron infections (89 of 143 [62.2%]) than for Delta infections (26 of 58 [53.0%]). Our findings of higher first-day sensitivity with Ag-RDT among participants infected with the Omicron variant may be attributable to these differences, which were not statistically significant.
In this study, more than half (52.7%) of the participants with a positive RT-PCR result had a false-negative result on an Ag-RDT even when 2 antigen tests were done within 48 hours of first RT-PCR positivity. However, when the analysis was restricted to participants who tested positive on RT-PCR for at least 48 hours (population B), the false-negative rate for Ag-RDT was 21.1% within 48 hours with no significant differences between the variant types. For the population of participants with singleton RT-PCR positivity, additional studies are needed to understand this phenomenon further in the context of SARS-CoV-2 infection compared with other viral infections where “blips” are commonly described (20–22). Such factors as SARS-CoV-2 immune status, local or systemic viral load, or assay limit of detection may play a role. The public health implications of false-negative Ag-RDT results associated with singleton RT-PCR positivity remain unclear (23). Because there is no way to prospectively determine who will remain positive on RT-PCR and who will have a singleton RT-PCR–positive result, it is important to elucidate the significance of our finding that Ag-RDTs fail to detect singleton RT-PCR–positive cases.
This analysis offers a unique look at longitudinal RT-PCR and Ag-RDT in a large prospective cohort, allowing us to capture data at the onset of infection and during the infection course throughout the emergence of the Omicron variant. This study used 3 different Ag-RDTs, which increases generalizability but does not guarantee it, and further evaluation of other Ag-RDTs may be needed as a clinical study. Identification of variants as Omicron or Delta in this study is based on sequencing of a subset of samples during December 2021 and the first week of January 2022, instead of all participants who tested positive. However, our observed sequencing results during December and January closely resemble those of the variant surveillance by the Centers for Disease Control and Prevention. To decrease possible misclassification of Delta and Omicron samples, we excluded participants with positive RT-PCR results but no sequencing results in the time when both Delta and Omicron were circulating. Furthermore, correction of possible misclassification error is unlikely to reverse the findings that Ag-RDTs have equal performance for the Delta and Omicron variants.
In conclusion, nasal swab Ag-RDT performance was similar between the Omicron and Delta variants. In both cases, detection of virus with Ag-RDTs was associated with relative viral load as measured by Ct value. Our data suggest that serial testing continues to be important in improving the performance of Ag-RDTs. Future work to increase our understanding of persons with singleton RT-PCR positivity is needed to determine the public health significance of a false-negative Ag-RDT result in this subpopulation.
Supplementary Material
Footnotes
This article was published at Annals.org on 11 October 2022.
References
- 1. Tromberg BJ , Schwetz TA , Pérez-Stable EJ , et al. Rapid scaling up of Covid-19 diagnostic testing in the United States — the NIH RADx initiative. N Engl J Med. 2020;383:1071-1077. [PMID: ] doi: 10.1056/NEJMsr2022263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mina MJ , Parker R , Larremore DB . Rethinking Covid-19 test sensitivity — a strategy for containment. N Engl J Med. 2020;383:e120. [PMID: ] doi: 10.1056/NEJMp2025631 [DOI] [PubMed] [Google Scholar]
- 3. The White House. Fact sheet: the Biden administration to begin distributing at-home, rapid COVID-19 tests to Americans for free. 14 January 2022. Accessed at www.whitehouse.gov/briefing-room/statements-releases/2022/01/14/fact-sheet-the-biden-administration-to-begin-distributing-at-home-rapid-covid-19-tests-to-americans-for-free on 28 August 2022.
- 4. Robinson ML, Mirza A, Gallagher N, et al. Limitations of molecular and antigen test performance for SARS-CoV-2 in symptomatic and asymptomatic COVID-19 contacts. medRxiv. Preprint posted online 7 February 2022. doi: 10.1101/2022.02.05.22270481 [DOI] [PMC free article] [PubMed]
- 5. Smith RL , Gibson LL , Martinez PP , et al. Longitudinal assessment of diagnostic test performance over the course of acute SARS-CoV-2 infection. J Infect Dis. 2021;224:976-982. [PMID: ] doi: 10.1093/infdis/jiab337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang Q , Syed AAS , Fahira A , et al. Structural analysis of the SARS-CoV-2 Omicron variant proteins. Research (Wash D C). 2021;2021:9769586. [PMID: ] doi: 10.34133/2021/9769586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boehm E , Kronig I , Neher RA , et al; Geneva Centre for Emerging Viral Diseases. Novel SARS-CoV-2 variants: the pandemics within the pandemic. Clin Microbiol Infect. 2021;27:1109-1117. [PMID: ] doi: 10.1016/j.cmi.2021.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu CR , Yin WC , Jiang Y , et al. Structure genomics of SARS-CoV-2 and its Omicron variant: drug design templates for COVID-19. Acta Pharmacol Sin. 2022. [PMID: ] doi: 10.1038/s41401-021-00851-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ferré VM , Peiffer-Smadja N , Visseaux B , et al. Omicron SARS-CoV-2 variant: what we know and what we don't [Editorial]. Anaesth Crit Care Pain Med. 2022;41:100998. [PMID: ] doi: 10.1016/j.accpm.2021.100998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Adamson BJ, Sikka R, Wyllie AL, et al. Discordant SARS-CoV-2 PCR and rapid antigen test results when infectious: a December 2021 occupational case series. medRxiv. Preprint posted online 5 January 2022. doi: 10.1101/2022.01.04.22268770 [DOI]
- 11. U.S. Food and Drug Administration. Omicron variant: impact on antigen diagnostic tests. In: SARS-CoV-2 Viral Mutations: Impact on COVID-19 Tests. Updated 28 December 2021. Accessed at www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests#omicronvariantimpact on 28 August 2022.
- 12. Kanjilal S, Chalise S, Shah AS, et al. Performance of three rapid antigen tests against the SARS-CoV-2 Omicron variant. medRxiv. Preprint posted online 19 February 2022. doi: 10.1101/2022.02.17.22271142 [DOI]
- 13. Deerain J , Druce J , Tran T , et al. Assessment of the analytical sensitivity of 10 lateral flow devices against the SARS-CoV-2 Omicron variant [Letter]. J Clin Microbiol. 2022;60:e0247921. [PMID: ] doi: 10.1128/jcm.02479-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stanley S, Hamel DJ, Wolf ID, et al. Limit of detection for rapid antigen testing of the SARS-CoV-2 Omicron variant. medRxiv. Preprint posted online 30 January 2022. doi: 10.1101/2022.01.28.22269968 [DOI]
- 15. Soni A, Herbert C, Pretz C, et al. Finding a needle in the haystack: design and implementation of a digital site-less clinical study of serial rapid antigen testing to identify asymptomatic SARS-CoV-2 infection. medRxiv. Preprint posted online 5 August 2022. doi: 10.1101/2022.08.04.22278274 [DOI] [PMC free article] [PubMed]
- 16. Soni A, Herbert C, Lin H, et al. Performance of screening for SARS-CoV-2 using rapid antigen tests to detect incidence of symptomatic and asymptomatic SARS-CoV-2 infection: findings from the Test Us at Home prospective cohort study. medRxiv. Preprint posted online 6 August 2022. doi: 10.1101/2022.08.05.22278466 [DOI]
- 17. Schrom J, Marquez C, Pilarowski G, et al. Direct comparison of SARS-CoV-2 nasal RT-PCR and rapid antigen test (BinaxNOW) at a community testing site during an Omicron surge. medRxiv. Preprint posted online 19 January 2022. doi: 10.1101/2022.01.08.22268954 [DOI]
- 18. Marais G, Hsiao NY, Iranzadeh A, et al. Saliva swabs are the preferred sample for Omicron detection. medRxiv. Preprint posted online 24 December 2021. doi: 10.1101/2021.12.22.21268246 [DOI]
- 19. Hay JA, Kissler SM, Fauver JR, et al. Viral dynamics and duration of PCR positivity of the SARS-CoV-2 Omicron variant. medRxiv. Preprint posted online 14 January 2022. doi: 10.1101/2022.01.13.22269257 [DOI]
- 20. Crowell TA , Pinyakorn S , Sacdalan C , et al; RV254/SEARCH010 Study Group. Viral blips after treatment initiation during acute human immunodeficiency virus infection. Clin Infect Dis. 2020;70:2706-2709. [PMID: ] doi: 10.1093/cid/ciz936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lodding IP , Mocroft A , da Cunha Bang C , et al. Impact of CMV PCR blips in recipients of solid organ and hematopoietic stem cell transplantation. Transplant Direct. 2018;4:e355. [PMID: ] doi: 10.1097/TXD.0000000000000787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brahmania M , Brouwer WP , Hansen T , et al. Prevalence and risk factors for viral blipping in chronic hepatitis B patients treated with nucleos (t) ide analogues. J Viral Hepat. 2016;23:1003-1008. [PMID: ] doi: 10.1111/jvh.12579 [DOI] [PubMed] [Google Scholar]
- 23. Liotti FM , Menchinelli G , Marchetti S , et al. Assessment of SARS-CoV-2 RNA test results among patients who recovered from COVID-19 with prior negative results [Letter]. JAMA Intern Med. 2021;181:702-704. [PMID: ] doi: 10.1001/jamainternmed.2020.7570 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.