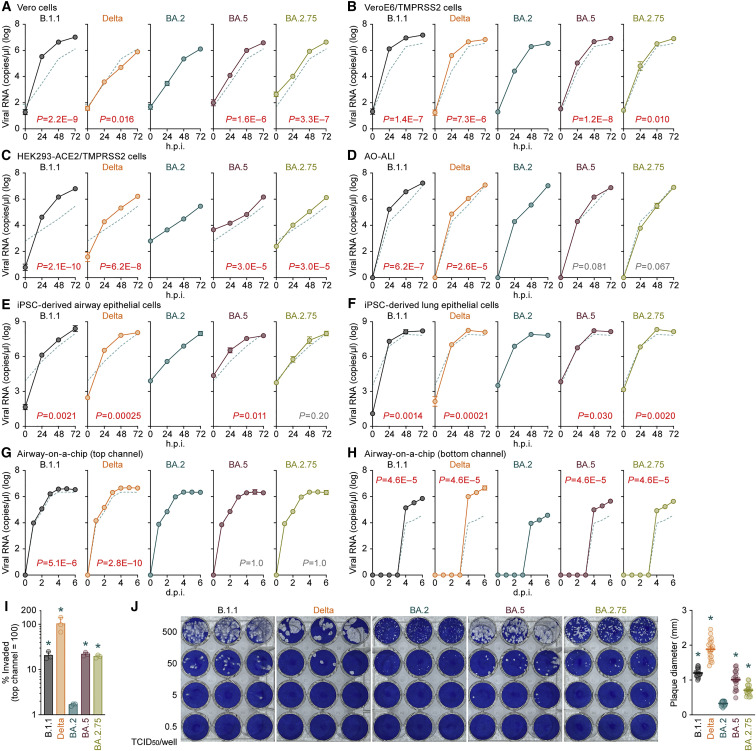
Figure 4.
Growth capacity of BA.2.75 in vitro
(A–I) Growth kinetics of B.1.1, Delta, BA.2, BA.5, and BA.2.75. Clinical isolates of B.1.1, Delta, BA.2, BA.5, and BA.2.75 were inoculated into Vero cells (A), VeroE6/TMPRSS2 cells (B), HEK293-ACE2/TMPRSS2 cells (C), AO-ALI (D), iPSC-derived airway epithelial cells (E), iPSC-derived lung epithelial cells (F), and an airway-on-a-chip system (G and H; see also Figure S3C). The copy numbers of viral RNA in the culture supernatant (A–C), the apical sides of cultures (D–F), and the top (G) and bottom (H) channels of an airway-on-a-chip were routinely quantified by RT-qPCR. The dashed green line in each panel indicates the results of BA.2. In (I), the percentage of viral RNA load in the bottom channel per top channel at 6 d.p.i. (i.e., % invaded virus from the top channel to the bottom channel) is shown.
(J) Plaque assay. Representative panels (left) and a summary of the recorded plaque diameters (20 plaques per virus) (right) are shown.
Assays were performed in quadruplicate, and the presented data are expressed as the average ± SD. In (A)–(H), statistically significant differences between BA.2 and the other variants across time points were determined by multiple regression. FWERs calculated using the Holm method are indicated in the figures. In (I) and (J) (right), statistically significant differences versus BA.2 (∗p < 0.05) were determined by two-sided Mann-Whitney U tests. Each dot indicates the result of an individual replicate.
See also Figure S4.