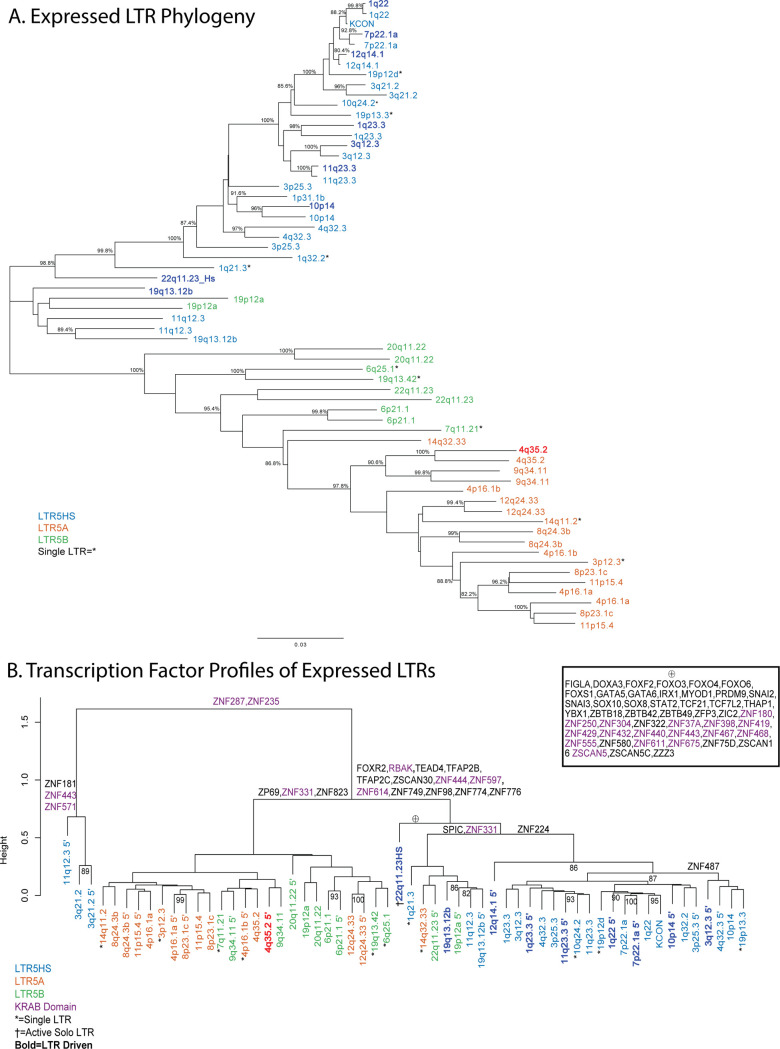
Fig 5. Relationships among multiple HML-2 proviruses of different LTR subtype.
(A) Neighbor-joining tree of the LTRs of each expressed HML-2 provirus in the GTEx dataset. Both LTRs are included when present, if only 1 LTR is present it is labeled with *. Color defines the subtype of each LTR as in Figs 1B and 3B. Darker color indicates proviruses whose expression is driven by the 5′ LTR. In both panels, KCON refers to the LTR of HERV-KCON, the infectious consensus HERV-K (HML-2) sequence [66]. (B) Cluster dendrogram of expressed HML-2 provirus LTRs based on TF binding profile as determined by FIMO (See Materials and methods). Solo LTRs are denoted by *. LTRs are colored by LTR5 subtype. A darker color and boldface signify an LTR observed to drive provirus expression. Defining TFs are shown above the branches of the dendrogram. Purple TFs are those that are known to have a KRAB domain [51]. The large number of TF motifs that define the branch containing the 22q11.23 LTR5HS solo LTR, which drives the expression of the adjacent LTR5B provirus [24], are shown in the inset under the ⊕ symbol. The sequences of HML-2 LTRs were collected from the HG38 human genomes using the coordinates included in S1 Table [8,10]. The matrix of TF sites is found in S3 Data. GTEx, Genotype Tissue and Expression; HML, human mouse mammary tumor virus-like; LTR, long terminal repeat; TF, transcription factor.