Abstract
Group3 (G3) medulloblastoma (MB) is one of the deadliest forms of the disease for which novel treatment is desperately needed. Here we evaluate ribociclib, a highly selective CDK4/6 inhibitor, with gemcitabine in mouse and human G3MBs. Ribociclib central nervous system (CNS) penetration was assessed by in vivo microdialysis and by IHC and gene expression studies and found to be CNS-penetrant. Tumors from mice treated with short term oral ribociclib displayed inhibited RB phosphorylation, downregulated E2F target genes, and decreased proliferation. Survival studies to determine the efficacy of ribociclib and gemcitabine combination were performed on mice intracranially implanted with luciferase-labeled mouse and human G3MBs. Treatment of mice with the combination of ribociclib and gemcitabine was well tolerated, slowed tumor progression and metastatic spread, and increased survival. Expression-based gene activity and cell state analysis investigated the effects of the combination after short- and long-term treatments. Molecular analysis of treated versus untreated tumors showed a significant decrease in the activity and expression of genes involved in cell-cycle progression and DNA damage response, and an increase in the activity and expression of genes implicated in neuronal identity and neuronal differentiation. Our findings in both mouse and human patient-derived orthotopic xenograft models suggest that ribociclib and gemcitabine combination therapy warrants further investigation as a treatment strategy for children with G3MB.
Introduction
Cyclin D-dependent kinases 4 and 6 (herein CDK4/6) and cyclin E/A-dependent CDK2 cooperatively catalyze the phosphorylation of the retinoblastoma tumor suppressor (RB) to inhibit its anti-proliferative function and facilitate entry into the DNA synthetic (S)-phase of the cell division cycle (1). Pathologic upregulation of proto-oncogenic D-type cyclins and CDK4/6, and genetic inactivation of specific polypeptide inhibitory proteins of CDK4/6 (INK4) are frequent events in many forms of cancer (1–3). Because stress-induced INK4 proteins, such as prototypic p16INK4a (encoded by CDKN2A) act to antagonize CDK4/6-mediated phosphorylation of RB, their ability to induce cell-cycle arrest prior to S-phase entry is strictly RB-dependent. Thus, cells lacking functional RB are resistant to CDK4/6 inhibition. Recently developed chemical inhibitors of CDK4/6, including palbociclib, ribociclib, and abemaciclib, that phenocopy the effects of INK4 proteins, have been FDA-approved for the treatment of estrogen receptor–positive breast cancer, and are currently in clinical trials for other cancers that retain functional RB (1).
Medulloblastoma (MB), one of the most frequent pediatric malignant brain tumors, is comprised of four consensus molecular subgroups: WNT, Sonic hedgehog (SHH), Group3 (G3), and Group4 (G4) (4). Among these subgroups, G3MB has the worst prognosis with frequent relapse, and survival ranging from only 40% to 60% despite conventional treatments, including surgery, radiation, and chemotherapy (5). MBs exhibit differential protein expression and recurrent copy-number aberrations of “RB pathway” genes, including CCND1, CCND2, CCND3, CDKN2A, CDKN2B, CDK4 and CDK6 (4, 6). Increased CDK6 protein expression is present in approximately 30% of MBs and is an independent poor prognostic marker of overall survival (7). These data suggest that targeting the RB pathway through the Cyclin D-CDK4/6 axis could be an effective way to suppress MB proliferation.
The three US Food and Drug Administration (FDA)-approved CDK4/6 inhibitors (CDK4/6i) induce G1 phase cell-cycle arrest and suppress tumor growth in preclinical models (1). Yet, the benefits from CDK4/6 inhibition are limited when given as monotherapy (8). Their failure, likely due to reversibility of cell cycle arrest upon drug withdrawal and to emergence of acquired drug resistance, can be bypassed by combining CDK4/6i with other agents to promote senescence or apoptosis (1). Gemcitabine, a metabolic inhibitor of DNA synthesis, is a lead candidate drug for G3 and G4MB and is currently in clinical trials in combination with pemetrexed (NCT01878617; SJMB12), and targeted agents [(NCT03434262; SJDAWN) and (NCT04023669; SJELIOT (9)]. In preclinical trials, administration of gemcitabine to mice implanted with patient-derived orthotopic xenografts (PDOX) and murine models of G3MB triggered tumor regression and significantly prolonged survival when combined with pemetrexed with or without standard of care (10). These effects, however, were transient and tumor regrowth on therapy suggested development of resistance. A reasonable argument might be made that CDK4/6i protect tumors from cytotoxic agents due to cell cycle exit (11) and antagonism has been reported with gemcitabine (12). However, an additive antitumor activity has also been described when combining CDK4/6i with gemcitabine (13), suggesting that there may be ways to effectively integrate these agents.
Recent data showed that ribociclib has good central nervous system (CNS) penetration and target modulation in human glioblastoma tissue samples (14, 15) and a preclinical study showed limited but sufficient CNS penetration in a mouse glioma model (16) to recommend evaluation of efficacy for treatment of CNS tumors.
Here, we assessed the pharmacokinetic and pharmacodynamic properties of the CDK4/6i ribociclib, an orally-bioavailable and highly selective CDK4/6-targeting agent, in preclinical models of G3MB to determine whether it is a viable therapeutic option for patients with this difficult-to-treat malignancy.
Materials and Methods
Cell lines and mouse and human G3MB models
MYC-amplified G3MB patient-derived orthotopic xenograft (PDOX) MB002, passage (p) 9–10, was generously provided by Yoon-Jae Cho (17) and SJMBG3-12-5950, p8, was previously described (18). Both G3MBs were amplified in Crl:CD1-Foxn1nu (CD1-nude) mice (Charles River Laboratories). Mouse G3MB tumor cell lines #2416 and #9730 were maintained as neurospheres, as previously published (19). Tumor cells were labeled with firefly luciferase (Luc2) to facilitate in vivo bioluminescence imaging (BLI), as previously reported (20). Tumors serially passaged in the cortices of naïve CD1-nude recipient mice were cryopreserved and banked for use in preclinical trials. Amplified PDOXs were molecularly characterized by Infinium Human Methylation 850K BeadChip (Illumina), IHC and FISH, and tested by DNA fingerprinting to ensure tumor identity. Mice were housed in an AAALAC-accredited facility. All animal studies were approved by the Animal Care and Use Committee and performed in accordance with best practices outlined by the NIH Office of Laboratory Animal Welfare.
Drug formulation, administration, and dose
Ribociclib succinate (Abmole) was prepared in 0.5% methylcellulose at 10 or 20 mg/mL free base equivalent and administered by oral gavage at 100 mg/kg or 200 mg/kg. 200 mg/kg dose was used for the survival study of mice bearing tumor #2416. Gemcitabine chloride (LC Laboratories) was prepared in sterile saline at 6 mg/mL free base equivalent and given by intravenous bolus at 60 mg/kg via tail vein injection.
Pharmacokinetic studies
To assess CNS penetration of ribociclib in CD1-nude mice bearing G3MB, we conducted plasma pharmacokinetic (PK) and cerebral microdialysis PK studies, as previously described (16).
Immunohistochemistry (IHC)
CD1-nude mice harboring MB002, SJMBG3-12-5950, or #2416 were treated with vehicle or ribociclib for 5 days. Treatment was initiated when BLI reached 1 × 107 or 1 × 108 photons/second (p/s) for mice bearing MB002 and SJMBG3-12-5950, respectively. For mice harboring tumor #2416, treatment was started 7 days post-implant [corresponding to 1 × 107–108 p/s]. Mice were sacrificed 4 hours after the last dose of ribociclib. Brains were fixed in 4% paraformaldehyde (PFA), embedded in paraffin, and sectioned (4 μm). Sections were deparaffinized, rehydrated, and submitted to antigen retrieval. Immunostaining of phospho-RBSer807/811 (Cell Signaling Technology, #CS8516, 1:100 dilution), activated Caspase 3 (BioCare Medical, #CP229C, 1:500 dilution) and Ki67 (ThermoShandon, #NuRM-9106, 1:50 dilution) used the Ventana Discovery Ultra XT (Ventana Medical). Heat-induced epitope retrieval used CC1 (Roche Medical). Antibodies were detected using OmniMap Rabbit (Roche Medical) followed by ChromeMap DAB (Roche Medical; ref. 21). Sections were counterstained with Hematoxylin II (Roche Medical). Slides were scanned on Aperio Imagescope system. An automatic count of positive cells was performed using nuclear V9 modified algorithm.
Preclinical trials
Luciferase positive tumor cells from #2416, #9730, MB002, passage (p) 9–10 [1 × 105 cells/mouse] or SJMBG3-12-5950, p8 [1 × 106 cells/mouse]) were resuspended in 5 μL Matrigel (BD Biosciences) and implanted in the cortices of 6- to 8-week old CD1-nude mice. Tumor growth was assessed twice weekly by BLI. Treatment was initiated 5 days post-implant for mouse models and when BLI was at least 5 × 105 p/s) for PDOXs. Treatment assignments were randomized. Ribociclib was administered daily for 3 weeks followed by 1 week off for mouse models. Because the ribociclib schedule administered to the mice bearing mouse G3MB models did not ultimately prevent tumor progression when combined with gemcitabine, subsequent studies with PDOXs used continuous daily ribociclib treatment hoping to improve survival. Gemcitabine was given every 2 weeks (starting on day 1). Treatment continued until mice were moribund. Control animals received vehicle (0.5% methylcellulose or saline) following the same schedule as ribociclib and gemcitabine, respectively. Study endpoint was defined by severe body weight loss (>20%), neurologic symptoms (seizure, abnormal gait), and/or poor body condition (lethargy, ataxia, severely hunched). Mice reaching these endpoints, were euthanized 4 hours post last dose. Tumors were harvested and snap frozen for molecular analysis.
Toxicity measurements
Toxicity was monitored via weekly blood work and body weights every two days. Complete blood counts from whole blood with EDTA were determined using a Forcyte Hematology Analyzer with impedance and laser technology (Oxford Scientific). Diagnostic chemistry panels were tested on sera by using the Trilogy chemistry analyzer (Drew Scientific).
Bioluminescence imaging
Tumor and spinal metastasis proliferation were assessed twice weekly by BLI, as previously published (18). Light intensity (p/s) was calculated using the LivingImage 4.5.2 software (PerkinElmer Inc.). Spinal images were acquired for mice bearing MB002 and line #9730 with the most pronounced metastases.
RNA sequencing, gene set enrichment analysis, and quantitative real-time PCR
Mice implanted with MB002, SJMBG3-12-5950, or #2416, were treated with vehicle, ribociclib, gemcitabine, or the combination. Tumors were harvested and snap frozen either 5 days post-enrollment or after long-term treatment (moribund stage). RNA was extracted using Trizol. After QC and library preparation, the library was sequenced using Illumina HiSeq platform. For detailed RNA sequencing methods and GSEA analysis, see Supplementary Data. qRT-PCR validation was performed, as previously described (22), with the exception of using the QuantStudio 3 Real-Time PCR system (Applied Biosystems) and the normalization by housekeeping genes GAPDH or B2m for human or mouse samples, respectively.
Reconstruction of medulloblastoma interactome and inference of gene activity
To reverse engineer a medulloblastoma-specific interactome (MBi), we applied SJARACNe (23) with default parameters (bootstrap = 100; consensus cluster P = 10–5; mutual information P = 10–7) to a medulloblastoma dataset (GSE85217; ref. 24) with expression profiles of 21,641 unique probe sets from 763 primary MB samples. See detailed description in Supplementary Data.
Gene activity was calculated using a cal.Activity function (es.method = ”weightedmean”, std = TRUE) in the NetBID software package (https://github.com/jyyulab/NetBID) based on the expression of their target genes from the MBi. For detailed description of the analysis, see Supplementary Data.
Expression of differentiation markers
Utilizing a 10× single-cell expression dataset of 78,156 cells from the developing mouse cerebellum spanning e10 to P10, differentiation scores were calculated for bulk RNA-seq cohorts (25). Nonnegative matrix factorization was applied to the centered mouse expression data, with negative values assigned to zero and rank set to two, to designate undifferentiated and differentiated programs. Both programs were projected onto a centered dataset of interest and the second component was determined as the differentiation score. G3 and G4MB differentiation markers were previously reported (25). Average expression enrichment of each gene set was determined using the score_genes function in the scanpy Python package for each sample in the bulk RNA-seq cohorts of interest. Comparison between treatment samples was calculated by Mann–Whitney test.
RNA-seq gene expression profiles for treatment samples were combined with those of 106 primary MB tumors for G3 and G4MB and corrected for batch using ComBat. Using genes with a standard deviation greater than 0.75, distances between samples were calculated by weighted Pearson correlation with SD as weights. The resulting distance matrix was visualized in two-dimensional space by implementing UMAP and plotting coordinates.
Statistical analysis
Overall survival (OS) was defined from study enrollment to euthanasia at predefined humane endpoints, as previously described (10). Statistical significance between treatment groups for IHC and relative mRNA quantity was determined by the Mann–Whitney test. For OS and treatment-related toxicity parameters, log-rank test (Mantel–Cox) adjusted for multiple comparisons (Holm–Sidak method) and mixed-effects models over time were used, respectively. GraphPad Prism version 7.0a for Mac (GraphPad Software, www.graphpad.com) was used for all analyses. Results were considered significant if P or adjusted P ≤0.05.
Data availability
RNA-seq data for MB002, #2416 and SJMBG3-12-5950 were deposited to the EGA (EGAS00001006001) and the St. Jude Cloud.
Results
Ribociclib penetrates CNS based on pharmacokinetic studies
CNS penetration of ribociclib in mice bearing #2416 was assessed after a single oral administration of 100 mg/kg ribociclib. Ribociclib disposition in plasma was best described using a one-compartment model with first-order absorption and first-order elimination, whereas, in tumor extracellular fluid (tECF) it was best described using a tumor compartment linked to a plasma compartment using influx and efflux clearances (Fig. 1A and B). The plasma exposure of ribociclib expressed as the area under concentration-time curve from time zero to 24 hours after dose (AUC0–24h) at 100 mg/kg dose in mice was comparable to that observed in humans at clinically relevant doses (i.e., 600 mg; ref. 26). The mean ± sd ribociclib tECF-to-plasma partition coefficient (Kp,uu; tECF AUC0–24h/unbound plasma AUC0–24h) was 0.162 ± 0.119, indicating that ribociclib crosses the blood brain barrier (BBB) in mice bearing G3MB (Table 1). These data are consistent with those obtained previously in non-tumor- and glioma-bearing mice (16).
Figure 1.
Ribociclib is brain penetrant and decreases pRB, Ki67, and the expression of E2F target genes in human and mouse G3MB tumors. A, Ribociclib plasma and extracellular fluid (ECF) model predictions in the mouse G3MB tumor #2416. CL, CL influx, CL efflux represents the elimination plasma to ECF influx and efflux clearances. OG, oral gavage; CL, clearance. B, Pharmacokinetic model structure. Unbound plasma concentrations (open black circles), mean model plasma predictions (solid black line), in tumor ECF concentrations (tECF, open red triangles), and mean model tECF predictions (dashed red line) in the mouse G3MB tumor #2416. Mice bearing human PDOX MB002 (C–F) or mouse tumor #2416 (G and H) tumors treated with vehicle or ribociclib (100 mg/kg), daily for 5 days and euthanized 4 hours post last dose. C and G, Representative IHC images tumors stained with antibodies against pRbSer807/811 (pRB), Ki67 and activated Caspase-3. D and H, Quantification of the percentage of positive cells for each staining [Mann–Whitney test, n = 7 per group for MB002 (D) and #2416, (H), P ≤ 0.05 (*), P ≤ 0.01 (**)]. Note: two untreated #2416 tumors added to #2416 vehicle group (H). E, Gene Set Enrichment Analysis (GSEA) showed significant depletion of HALLMARK_E2F_TARGETS in ribociclib versus vehicle-treated MB002 tumors. F, Relative mRNA level of selected E2F target genes quantified by qRT-PCR [Mann–Whitney test; n = 4 per group; P ≤ 0.05 (*)].
Table 1.
Ribociclib pharmacokinetic parameters summary in mice bearing mouse tumor #2416 (n = 7).
| Parameters [unit] | Mean estimates (SD) |
|---|---|
| Half-life (h) | 4.26 (0.737) |
| Unbound Plasma AUC0–24h (ng × h/mL) | 5,820 (926) |
| tECF AUC0–24h/unbound Plasma AUC0–24h | 0.162 (0.119) |
Abbreviations: AUC0–24h, area under concentration–time curve from time zero to 24 hours after dose; tECF, tumor extracellular fluid.
Ribociclib decreases RB phosphorylation and represses the transcription of E2F target genes
We then tested ribociclib pharmacodynamic properties in tumors after a short-term treatment of mice harboring G3MB human MB002 (Fig. 1C–F) or mouse #2416 (Fig. 1G and H). Mice were treated daily with 100 mg/kg orally of ribociclib or vehicle for 5 days. Tumors were harvested 4 hours after the last dose. In MB002 tumors, we found a significant reduction of phosphorylated RB (pRBser807/811) and proliferation as measured by Ki67 staining for mice treated with ribociclib compared with vehicle control (Fig. 1C and D). As expected from a cytostatic drug, we did not detect significant cell death, measured by activated caspase 3 staining (Fig. 1C and D). GSEA revealed that E2F target genes were depleted in ribociclib versus vehicle treated tumors (Fig. 1E). Downregulation of several of these genes, CCNE2, CCNA2, MKI67, TOP2A and PLK1, was confirmed by qRT-PCR (Fig. 1F).
Similar results were obtained with mouse tumor #2416 (Fig. 1G and H) and PDOX SJMBG3–12–5950 (Supplementary Fig. S1A–S1D) although the decrease of pRB and Ki67 positive cells did not reach significance. The depletion of E2F target genes after ribociclib treatment was not statistically significant in the #2416 cohort (Supplementary Fig. S2A) notably owing to two samples from mice that had higher luminescence signal at treatment initiation and failed to respond (Supplementary Fig. S2B–S2D; Supplementary Table S1), possibly due to the relatively high tumor burden.
Altogether, these data demonstrate on-target effects of ribociclib in vivo in mouse and human G3MBs, with variation in the extent of target inhibition between models after 5 days of treatment.
Combination treatment with ribociclib and gemcitabine is well tolerated and significantly prolongs survival
We conducted survival studies in mice bearing intracranial G3MBs to investigate the efficacy of ribociclib alone and in combination with gemcitabine. We previously showed that treatment of mice bearing G3MBs with gemcitabine, a nucleoside analog, prolonged their survival when combined with pemetrexed (10). To investigate whether combining ribociclib with gemcitabine would result in survival improvement, we used two ribociclib schedules. Mice bearing PDOX (MB002 and SJMBG3-12-5950) were treated with vehicle, 100 mg/kg of ribociclib once daily, a dose that compares to a previously reported phase I study (26), gemcitabine 60 mg/kg once every two weeks, or the two drugs in combination until moribund (Fig. 2A). Mice bearing mouse tumors #2416 and #9730 were treated with vehicle, 100 mg/kg of ribociclib or 200 mg/kg, a dose widely used by others (27), once daily for three weeks followed by one week off, gemcitabine 60 mg/kg once every two weeks, or the two drugs in combination with continuous cycles (Fig. 2B).
Figure 2.
Combination therapy of ribociclib with gemcitabine improves survival of mice bearing G3MB tumors. A and B, Treatment schedules for human and mouse models, respectively. C–G, Mice bearing PDOX MB002 tumor cells treated with vehicle (black lines), ribociclib [100 mg/kg, continuous daily by oral gavage (OG), red lines], gemcitabine (60 mg/kg, i.v., day1 then every 2 weeks, blue lines), or the combination (purple lines) until moribund. H–K, Mice bearing mouse tumor #2416 treated with vehicle (black lines), ribociclib [100 or 200 mg/kg, day1–21 (3 weeks), OG, red lines], gemcitabine (60 mg/kg, i.v., day 1 then every 2 weeks, blue lines), or the combination of gemcitabine (60 mg/kg, i.v., day1 then every 2 weeks) with ribociclib (100 or 200 mg/kg, day1–21, OG, purple lines). Mice were treated with sequential cycles of therapy until moribund. C and H, Kaplan–Meier survival plots for all treatment groups. Comparison between treatment groups using log-rank test adjusted for multiple comparisons [not significant (n.s.), adjusted P ≤ 0.05 (*), adjusted P ≤ 0.005 (**), and adjusted P ≤ 0.0005 (***)]. Mice were censored if they died or required humane euthanasia not related to tumor and without preceding neurologic symptoms (e.g., death during sedation for imaging). Mice were imaged twice weekly by bioluminescence imaging (BLI). D and I, BLI pictures at different time points from enrollment through moribund stage represented for one mouse per treatment group. Mice were selected based on tumor and spinal growth median behavior. E, F, and J, BLI increase from enrollment (LOG10 transformation) for brain (E and J) and spine [F; Mann–Whitney test, P ≤ 0.05 (*), P ≤ 0.01 (**)]. For all BLI signal curves, see Supplementary Fig. S3. G and K, Endpoint BLI images of mice with visible spinal signal for MB002 (G), and #2416 (K), based on respective scales. Please note that the endpoint images of the vehicle and ribociclib mice presented in D were the same as in G where they are represented as part of all the mice imaged at endpoint in the study.
In mice bearing MB002 (Fig. 2C–G), ribociclib alone significantly prolonged survival compared with control with a 10.5 day median survival advantage (Fig. 2C; median survival: 37 for the ribociclib versus 26.5 days for the vehicle treated mice; adjusted P = 0.0015). Combination of ribociclib and gemcitabine further improved survival by 11 days compared with mice treated with ribociclib alone with a 21.5 day difference when compared with vehicle treated mice [Fig. 2C; median survival: 48 days for the combination treated mice; adjusted P = 0.0015 (compared with ribociclib) and adjusted P = 0.0005 (compared with vehicle)]. Similarly, in SJMBG3–12–5950-bearing mice (Supplementary Fig. S3A–S3D), the combination prolonged survival compared to vehicle (Supplementary Fig. S3A; median survival: 63.5 for the combination versus 46.5 days for the vehicle treated mice; adjusted P = 0.0218).
In mice bearing mouse MB #2416 (Fig. 2H–K), no difference of survival was observed between 100 and 200 mg/kg doses of ribociclib (Supplementary Fig. S4), therefore we combined the mice treated with the two different doses (Fig. 2H). In this model, the combination of ribociclib and gemcitabine significantly prolonged survival when compared with controls whereas single agents did not. Median survival for mice treated with the combination versus control was 35 versus 15 days (Fig. 2H; adjusted p value = 0.0028). In mice bearing the mouse line #9730 (Supplementary Fig. S5A–S5E), the combination of ribociclib and gemcitabine did not significantly prolong survival when compared to controls. Median survival for mice treated with the combination versus control was 47 versus 39.5 days (Supplementary Fig. S5A; adjusted P = 0.0625). Gemcitabine as single agent had little to no effect on the survival of G3MB-bearing mice in all four models, (Fig. 2C and H; Supplementary Figs. S3A and S5A). All mice included in the survival studies were imaged twice weekly; see BLI for the brain (Supplementary Fig. S6A–S6D and S6I-T) and the spine (Supplementary Fig. S6E–S6H and S6U–S6X).
Treatment-related toxicity profiles were similar for all animals even under continuous ribociclib treatment. We observed only mild leukopenia in ribociclib and combination arms but no severe neutropenia (Supplementary Fig. S7A and S7B). We also did not detect significant treatment-related anemia (Supplementary Fig. S7C), thrombocytopenia (Supplementary Fig. S7D), elevations in liver transaminases AST and ALT (Supplementary Fig. S7E and S7F, respectively) or kidney function (Supplementary Fig. S7G). Decreases in body weight correlated with tumor progression accross treatment arms with no indication of treatment-related weight loss (Supplementary Fig. S7H).
Combination treatment with ribociclib and gemcitabine delays tumor progression and metastasis spread
The survival improvement of mice treated with the combination of ribociclib and gemcitabine was associated with delayed tumor growth in the brain and delayed onset of spinal metastases (Fig. 2D–F, I and J; Supplementary Figs. S3B, S3C, S5B-S5D). Eventually, all mice succumbed to disease. However, we found a difference in the extent of metastases between models at the moribund stage: MB002- and #9730-bearing mice showed higher BLI signal in the spine, 33/40 (82%) and 17/28 (61%), respectively (Fig. 2G; Supplementary Fig. S5E), than in mice bearing SJMBG3-12-5950 [10/31 (32%)] and #2416 [6/33 (18%)] (Supplementary Fig. S3D; Fig. 2K).
Combination of ribociclib and gemcitabine decreases the activity of cell cycle drivers and genes involved in DNA damage response
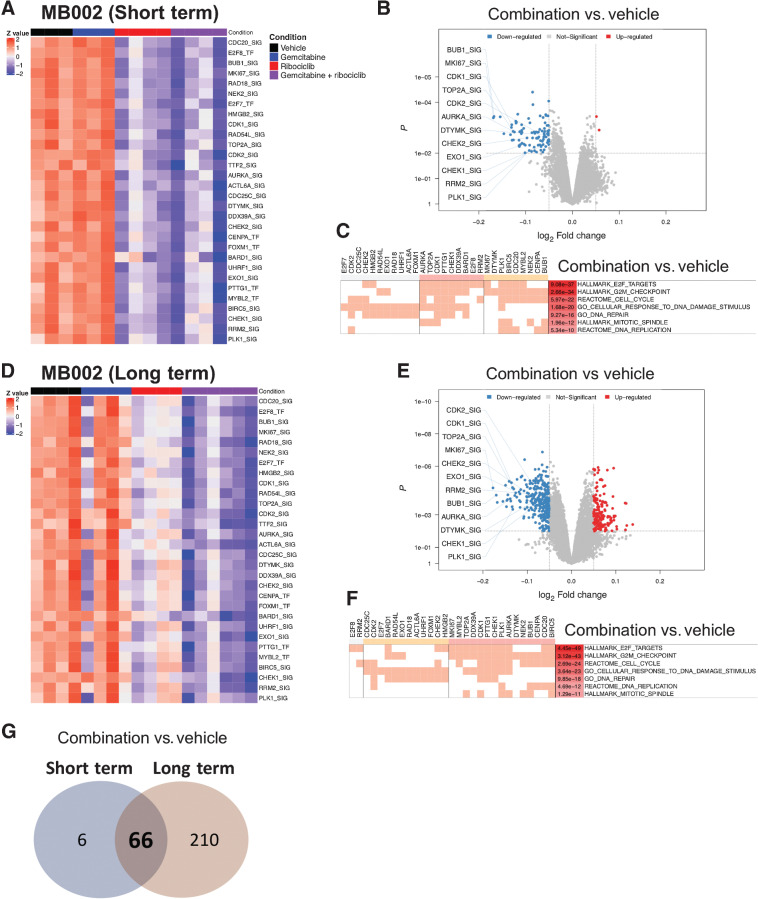
To identify putative ribociclib and gemcitabine transcriptional targets, we performed RNA-seq profiling of tumors harvested from MB002-bearing mice treated with vehicle, gemcitabine, ribociclib, or gemcitabine and ribociclib for 5 days (Fig. 3A–C; Supplementary Fig. S8A) or at moribund stage (Fig. 3D–F). We used the NetBID algorithm (28) to infer driver gene activity from the RNA-seq data and a MB-specific gene network.
Figure 3.
Combination of ribociclib and gemcitabine decreases the activity of genes involved in cell cycle regulation and DNA damage response in MB002 tumors. Mice bearing human MB002 PDOX treated with vehicle (black bar), gemcitabine (60 mg/kg, blue bar), ribociclib (100 mg/kg, red bar) or both (purple bar). Tumors were harvested on day 5 after short term treatment (A–C) or after long-term treatment at moribund stage (D–F). A and D, NetBID analysis to infer gene activity from RNA-seq data and a MB-specific interactome. Activity heatmaps of a selection of genes in tumors from the four treatment groups. B and E, Volcano plots showing genes with differential activity between tumors treated with gemcitabine and ribociclib versus vehicle (network size > 25; P < 0.01 and logFC < -0.05 or logFC > 0.05). C and F, Clustering of genes involved in down-regulated pathways. G, Venn diagram showing the number of differentially expressed genes from tumors treated with gemcitabine and ribociclib versus vehicle between short-term and long-term treatments.
After short term treatment, based on NetBID, 72 genes were differentially activated between tumors treated with the combination versus vehicle. Among these, 30 genes regulated the cell cycle and DNA damage response (Fig. 3C). Gemcitabine treatment, when given on day 1 and tumors harvested on day 5, had no effect on the activity profile of these genes (Fig. 3A). To confirm that gemcitabine induced DNA damage in vivo, tumors were immunostained for γH2AX. As expected, when tumors were harvested 4 hours after treatment (acute treatment), gemcitabine increased the number of γH2AX-positive cells (Supplementary Fig. S8A–S8C). However, 4 days after gemcitabine treatment, the percentage of γH2AX-positive cells was identical to that of vehicle-treated tumors (Supplementary Fig. S8A–S8C). The activity of the 30 genes were similarly decreased in tumors treated with ribociclib or the ribociclib and gemcitabine combination versus vehicle after short-term treatment (Fig. 3A and B). GSEA of tumors treated with the combination of gemcitabine and ribociclib compared with vehicle revealed significant depletion of the activity of genes that regulate the cell cycle and the DNA damage response including E2F targets, G2M checkpoint, DNA replication, mitotic spindle, DNA damage, and DNA repair (Fig. 3C).
Long-term treatment with combination of ribociclib and gemcitabine resulted in a more pronounced downregulation of the activity profile of the 30 genes compared with treatment with ribociclib alone (Fig. 3D). Similar to short-term treatment, the activity of the 30 genes was significantly downregulated (Fig. 3E), and the same classes of genes were depleted when comparing combination versus vehicle treated tumors (Fig. 3F). Overall, 66 genes were differentially activated in both short- and long-term studies (Fig. 3G).
After a short-term or long-term treatment with the drug combination, decreased gene activity seen in tumors was associated with a decrease in RNA expression (Supplementary Fig. S8D and S8E) confirmed by qRT-PCR (Supplementary Fig. S8F and S8G). Genes with downregulated activity and expression included many E2F targets including E2F8, MKI67, PLK1, RRM2, and TOP2A. Data obtained in the MB002 model were similar in long-term treated SJMBG3-12-5950 and #2416 tumor models (Supplementary Fig. S9A–S9F).
Overall, these results suggest that gemcitabine and ribociclib combination is effective in decreasing the activity of regulators of cell-cycle and DNA damage response after short-term and long-term treatments.
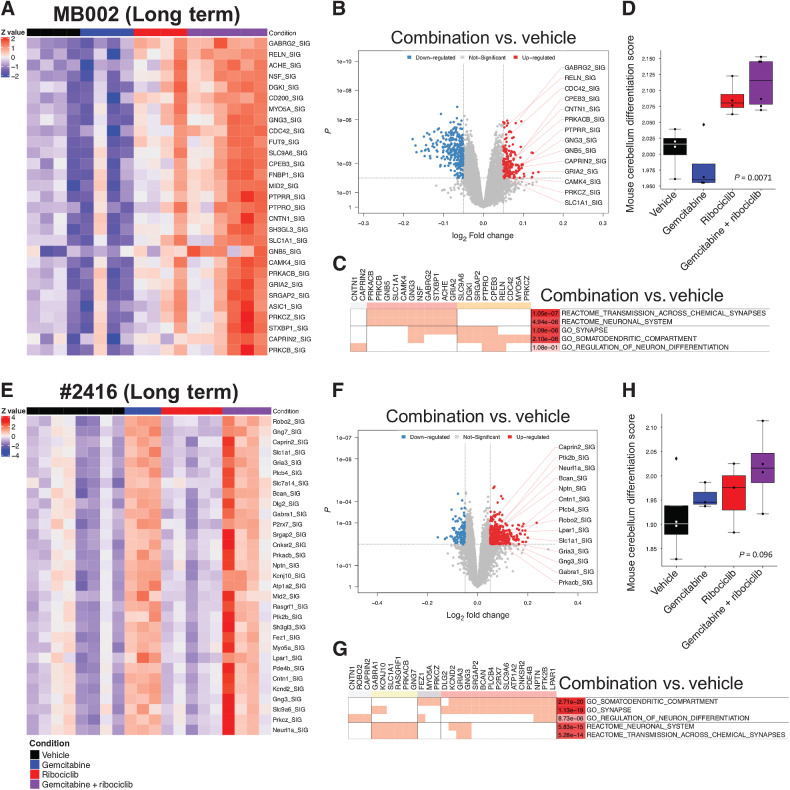
Long-term treatment with the combination of ribociclib and gemcitabine increases the activity of genes involved in neuronal identity and differentiation
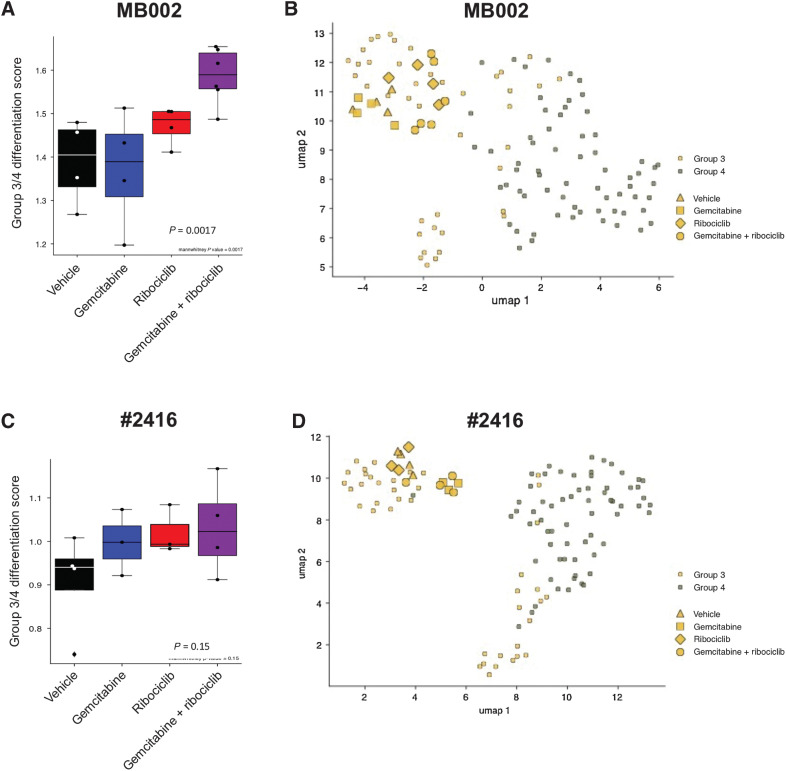
After long-term treatment with the drug combination in MB002 (Fig. 4A–D), #2416 (Fig. 4E–H), and SJMBG3-12-5950 (Supplementary Fig. S10A–S10C), NetBID activity and GSEA analyses uncovered activity signatures for neuronal identity and neuronal differentiation. For MB002 and SJMBG3-12-5950, the increased activity was mainly seen in tumors treated with the combination, whereas this effect was also seen in gemcitabine-treated #2416 tumors (Fig. 4A and E; Supplementary Fig. S10A). All selected genes were significantly activated in tumors treated with gemcitabine and ribociclib versus vehicle (Fig. 4B and F; Supplementary Fig. S10B). In MB002 and #2416 tumors, similar gene sets were enriched in tumors treated with the drug combination compared to vehicle. This included classes of genes associated with neuronal identity, synaptogenesis, and neuronal differentiation (Fig. 4C and G). In all three models, we found significantly activated genes belonging to these gene sets (Fig. 4B, C, F, G; Supplementary Fig. S10B). However, we failed to see enrichment of these pathways in SJMBG3-12-5950 tumors treated with the drug combination. Using a single-cell RNA-seq dataset from the developing mouse cerebellum (29), differentiation scores were computed for each tumor. In all models, tumors treated with ribociclib and gemcitabine displayed a higher neuronal differentiation score than vehicle treated tumors (Fig. 4D and H; Supplementary Fig. S10C). Using cell states defined in a recent single-cell RNA-seq study of MB (25), expression of genes included in the G3/G4 differentiation program were also increased in G3MB tumors treated with the combination (Fig. 5A and C; Supplementary Fig. S10D), although, they still clustered within the G3 subgroup (Fig. 5B and D; Supplementary Fig. S10E).
Figure 4.
Long-term combination of ribociclib and gemcitabine increases the activity of genes involved in neuronal identity in G3MB. NetBID analysis and differentiation score of tumors from mice bearing PDOX MB002 (A–D) or mouse tumor #2416 (E–H) treated with vehicle (black bar), gemcitabine (blue bar), ribociclib (red bar), and gemcitabine and ribociclib (combination) (purple line). A and E, Activity heatmaps of a selection of genes from tumors for each treatment group. B and F, Volcano plots showing genes with differential activity in tumors treated with gemcitabine and ribociclib combination versus vehicle (network size > 25; P < 0.01 and logFC < -0.05 or logFC > 0.05). C and G, Clustering of genes in upregulated pathways. D and H, Differentiation scores for tumors for each treatment group computed from a single-cell RNA-seq dataset from the developing mouse cerebellum (Mann–Whitney test, gemcitabine + ribociclib versus vehicle).
Figure 5.
G3MB tumors treated with the combination of ribociclib and gemcitabine have a higher differentiation score and remain of the G3 subgroup. Analysis of the expression of genes in the G3/G4 differentiation pathways compared to those expressed in primary human MB tumors after RNA sequencing from PDOX MB002 (A and B) or mouse tumor #2416 (C and D) after treatment with vehicle, gemcitabine, ribociclib and combination of gemcitabine and ribociclib. A and C, G3/G4 differentiation score (Mann–Whitney test, gemcitabine + ribociclib versus vehicle). B and D, Clustering of the tumors with primary human G3 and G4 MB tumors.
Discussion
While the majority of children with medulloblastoma are cured with conventional therapy, about one-quarter succumb to the disease. Among these failures, patients with G3MB are over-represented necessitating better therapy (30, 31). Our study adds to a growing body of evidence that CDK4/6i impair proliferation of MB cells (32–34) and that ribociclib is CNS penetrant, albeit modestly (14–16). We show that the ribociclib/gemcitabine combination results in significantly improved survival in mice bearing intracranially implanted mouse and human G3MB tumors. Combination therapy was well-tolerated, and toxicity was limited to leukopenia without severe neutropenia similar to treatment with ribociclib alone. A recent phase I trial of ribociclib in children demonstrated acceptable safety and established a recommended phase II dose equivalent to adults (35), and gemcitabine has been well tolerated when combined with other agents in early phase studies in children (36, 37). The ribociclib/gemcitabine combination in children may be limited by hematologic toxicity based on prior experience with CDK4/6i in children as single agents (8, 35). However, we are encouraged that achievable levels of the agents at tolerable doses may be sufficient to induce meaningful clinical responses despite the limited CNS exposure compared to systemic exposure. The difference seen in the response to treatment between the models tested might mirror what is seen in patients with relapsed G3MB. Because relapsed medulloblastoma is a fatal disease with limited treatment options, results obtained in the best available model systems are used for exploring this potential therapy. The identification of reliable biomarkers that predict the response to treatment is also warranted.
Molecular characterization of tumors treated with ribociclib and gemcitabine
The combination of ribociclib and gemcitabine was associated with the inhibition of activity and gene expression of cell-cycle genes, including E2F targets, and of genes in the DNA damage signaling and repair pathways. These data are consistent with multiple in vitro and in vivo studies in which CDK4/6 inhibition or CDK6 knock-down was combined with DNA-damaging agents and was associated with an increase of DNA damage assessed by γH2AX staining (38). Thus, we hypothesize that ribociclib, by blocking E2F activity, is preventing tumor cell recovery by impairing the DNA damage signaling and activity of repair genes induced following gemcitabine treatment. The activity of this combination in MYC-driven G3MBs could be explained by the high level of replicative stress and susceptibility to targeted therapies inhibiting DNA repair (39, 40). Interestingly, in human MBs proteomic data, G3MBs expressed high protein level and phosphoprotein signature of DNA repair pathways (41, 42). Consistently, CHK1/2 inhibition was found to potentiate gemcitabine activity in vivo in mice bearing G3MB (9). Furthermore, by decreasing the activity and expression of the ribonucleotide reductase E2F target genes RRM1 and RRM2, ribociclib may cooperate with gemcitabine to delay tumor progression by affecting ribonucleotide synthesis (13, 43).
Finally, we showed that when mouse and human G3MBs are treated with the combination of ribociclib and gemcitabine, genes involved in cerebellar and neuronal differentiation were upregulated and activated. While MYC-amplified G3MBs are characterized by undifferentiated stem-like neuronal cells and G4MBs are predominantly comprised of more differentiated neuronal cells, a proportion of MBs are intermediate between clear G3 and G4MBs (25). Although combination treatment of G3MB with ribociclib and gemcitabine promoted a neuronal expression profile, G3 subgroup status was maintained in these tumors, indicating that these tumors were shifted towards a more differentiated phenotype rather than a different subgroup.
Combination therapy was beneficial but not curative
Although the combination of ribociclib and gemcitabine provided mice bearing mouse or human G3MB with a survival advantage, all mice ultimately succumbed to tumor burden. Molecular analysis of tumors after short or long term treatment suggests that the survival advantage might be due to cytostasis and pro-differentiation leading to reduced tumor proliferation. It is possible that the administration schedule we used, based on prior clinical experience to facilitate easy translation into human trials, was not optimal. There is a rationale to use CDK4/6i to synchronize and thereby sensitize cells to subsequent cytotoxic therapy (44). Several studies suggest that co-treatment of CDK4/6i with chemotherapy or the use of chemotherapy before CDK4/6 inhibition is a promising strategy (38). Therefore, exploration of different timing/sequencing of ribociclib and gemcitabine as well as ways to incorporate this regimen into more standard MB therapy remain a priority.
In addition, while we found a modest degree of CNS penetration of ribocilib and observed differences in key biomarkers in tumor tissue, antitumor efficacy may have been limited by relatively lower CNS exposure at tolerable systemic doses, an ongoing challenge in the treatment on CNS tumors. Indeed, in a subcutaneous medulloblastoma PDX model a greater difference in Ki67 positivity was observed in mice treated with the CDK4/6i palbociclib (33) compared with our results. Far more suppression of Ki67 labeling was seen in specimens from patients with breast cancer treated with abemacilib and ribociclib (26, 45) suggesting a dose-dependent on-target effect based on tissue exposure.
Other combinations strategies
To further improve survival and possibly induce a more durable remission, additional agents that could be combined with ribociclib should also be considered. A recent study showed that the addition of docetaxel to gemcitabine and ribociclib or palbociclib blocked tumor growth of PDX models of pancreatic ductal adenocarcinoma (46). Another possible avenue includes reinforcing CDK4/6 inhibition with D-type cyclin inhibition. Induction of the D-type cyclins is known to occur from a number of signal transduction pathways such as the RTK-RAS-MEK-ERK, SHH, WNT, NF-kB and PI3K that are also commonly deregulated in cancer and often in conjunction with lesions in the cyclinD-Rb-E2F pathway (2). Dual or triple pharmacological blockade of these signal transduction pathways in conjunction with CDK4/6i may be required for sustained responses. For example, synergy has been described between inhibitors of MEK, PI3K/mTOR and CDK4/6 in multiple preclinical studies (38, 47). Interestingly, senescence and vascular remodeling associated with CDK4/6 and MEK inhibition was shown to sensitize the KRAS mutant pancreatic ductal adenocarcinoma to gemcitabine (48).
Our findings suggest that the combination of gemcitabine with a different CDK4/6i could also be efficacious if CNS penetration is sufficient at tolerable systemic doses. A recently published physiologically-based pharmacokinetic modeling approach demonstrated predicted target engagement ratios (ratio of the average steady-state unbound drug brain concentration to the in vitro IC50 for CDK4/6 inhibition) following standard dosing regimens for CDK4/6 of 26/5.2 for abemaciclib, 2.4/0.62 for ribociclib, and 0.36/0.27 for palbociclib concluding that abemaciclib may be the most efficacious for brain cancer treatment (49).
Alternatively, the combination of gemcitabine with CHK1/2 inhibitors dramatically improved the survival of mice bearing G3MBs (9) and is currently evaluated in the clinic (SJELIOT: NCT04023669).
CDK4/6i for the treatment of all MB subgroups
We focused on G3MB, the subgroup with one of the worst prognoses (5). However, CDK4/6i therapy may be useful in the other MB subgroups. Preclinical in vivo efficacy in SHH MB was observed with palbociclib (33) and G4MBs display focal CDK6 amplification in 5% of tumors suggesting some targetable activity (4). Herein, we incorporated gemcitabine into treatment given previously observed activity against G3MB. However, other agents such as SHH inhibitors or alternative chemotherapies could and should be explored for the other subgroups.
Conclusion
Our data confirm ribociclib as a CNS penetrant CDK4/6i and provide rationale for combining ribociclib and gemcitabine for the treatment of G3MB, but further investigation is warranted. These findings helped form the basis for a phase I clinical trial (NCT03434262) in patients with recurrent G3 or G4MB who relapsed after conventional therapy. Data from the aforementioned clinical trial as well as additional preclinical research are needed to determine the most effective ways to combine CDK4/6i with other agents for the treatment of children with medulloblastoma and other CNS tumors.
Supplementary Material
Acknowledgments
This research was supported by NIH grants CA-096832 and CA-02165 (MFR) and the American Lebanese-Syrian Associated Charity of St. Jude Children's Research Hospital. We thank members of the Medulloblastoma Working Group and the Roussel/Sherr laboratory for helpful discussions. We are indebted to Justin S. Williams for computational analysis; Frederique Zindy, Dana Farmer, Jose Grenet and Kimberly Shea Mercer for technical expertise; Center for In Vivo Imaging and Therapeutics; Animal Resource Center technical services; Veterinary Pathology Core; Core Flow Cytometry and Cell Sorting and Hartwell center. We thank Yoon-Jae Cho for MB002 cells. Charles J. Sherr helped in formulating the initial manuscript.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This article is featured in Highlights of This Issue, p. 1259
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Disclosures
M.F. Roussel reports grants from NIH Grant CA-096832, grants from NIH Grant CA-02165, and other support from ALSAC during the conduct of the study; personal fees from CSHL, personal fees from Fred Hutchinson Cancer Research Center, personal fees from John's Coffin Childs, and personal fees from AACR MCT outside the submitted work. No disclosures were reported by the other authors.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Authors' Contributions
A. Pribnow: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. B. Jonchere: Data curation, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. J. Liu: Software, validation, visualization, methodology, writing–review and editing. K.S. Smith: Data curation, software, validation, investigation, visualization, methodology, writing–review and editing. O. Campagne: Data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. K. Xu: Data curation, software, formal analysis, methodology. S. Robinson: Formal analysis, investigation, visualization, methodology, writing–review and editing. Y. Patel: Investigation. A. Onar-Thomas: Data curation, formal analysis, validation, investigation, methodology, writing–review and editing. G. Wu: Data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. C.F. Stewart: Data curation, formal analysis, validation, investigation, methodology, writing–review and editing. P.A. Northcott: Data curation, software, formal analysis, investigation, visualization, methodology, writing–review and editing. J. Yu: Data curation, software, formal analysis, investigation, visualization, methodology, writing–review and editing. G.W. Robinson: Data curation, formal analysis, validation, investigation, methodology, writing–review and editing. M.F. Roussel: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, writing–original draft, project administration, writing–review and editing.
References
- 1. Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: from discovery to therapy. Cancer Discov 2016;6:353–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sherr CJ. Cancer cell cycles. Science 1996;274:1672–7. [DOI] [PubMed] [Google Scholar]
- 3. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57–70. [DOI] [PubMed] [Google Scholar]
- 4. Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 2012;488:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kool M, Korshunov A, Remke M, Jones DTW, Schlanstein M, Northcott PA, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, group 3, and group 4 medulloblastomas. Acta Neuropathol 2012;123:473–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Uziel T, Zindy F, Xie S, Lee Y, Forget A, Magdaleno S, et al. The tumor suppressors Ink4c and p53 collaborate independently with Patched to suppress medulloblastoma formation. Genes Dev 2005;19:2656–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mendrzyk F, Radlwimmer B, Joos S, Kokocinski F, Benner A, Stange DE, et al. Genomic and protein expression profiling identifies CDK6 as novel independent prognostic marker in medulloblastoma. J Clin Oncol 2005;23:8853–62. [DOI] [PubMed] [Google Scholar]
- 8. Van Mater D, Gururangan S, Becher O, Campagne O, Leary S, Phillips JJ, et al. A phase I trial of the CDK 4/6 inhibitor palbociclib in pediatric patients with progressive brain tumors: a pediatric brain tumor consortium study (PBTC-042). Pediatr Blood Cancer 2021;68:e28879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Endersby R, Whitehouse J, Pribnow A, Kuchibhotla M, Hii H, Carline B, et al. Small-molecule screen reveals synergy of cell cycle checkpoint kinase inhibitors with DNA-damaging chemotherapies in medulloblastoma. Sci Transl Med 2021;13:eaba7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morfouace M, Shelat A, Jacus M, Freeman BB, 3rd, Turner D, Robinson S, et al. Pemetrexed and gemcitabine as combination therapy for the treatment of Group3 medulloblastoma. Cancer Cell 2014;25:516–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McClendon AK, Dean JL, Rivadeneira DB, Yu JE, Reed CA, Gao E, et al. CDK4/6 inhibition antagonizes the cytotoxic response to anthracycline therapy. Cell Cycle 2012;11:2747–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Franco J, Witkiewicz AK, Knudsen ES. CDK4/6 inhibitors have potent activity in combination with pathway selective therapeutic agents in models of pancreatic cancer. Oncotarget 2014;5:6512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gelbert LM, C S, Lin X, Sanchez-Martinez C, Del Prado M, Lallena MJ, et al. Preclinical characterization of the CDK4/6 inhibitor LY2835219: in-vivo cell cycle-dependent/independent anti-tumor activities alone/in combination with gemcitabine. Invest New Drugs 2014;32:825–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tien AC, Li J, Bao X, Derogatis A, Kim S, Mehta S, et al. A phase 0 trial of ribociclib in recurrent glioblastoma patients incorporating a tumor pharmacodynamic- and pharmacokinetic-guided expansion cohort. Clin Cancer Res 2019;25:5777–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bao X, Wu J, Sanai N, Li J. Determination of total and unbound ribociclib in human plasma and brain tumor tissues using liquid chromatography coupled with tandem mass spectrometry. J Pharm Biomed Anal 2019;166:197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Patel YT, Davis A, Baker SJ, Campagne O, Stewart CF. CNS penetration of the CDK4/6 inhibitor ribociclib in non-tumor bearing mice and mice bearing pediatric brain tumors. Cancer Chemother Pharmacol 2019;84:447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bandopadhayay P, Bergthold G, Nguyen B, Schubert S, Gholamin S, Tang Y, et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin Cancer Res 2014;20:912–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smith KS, Xu K, Mercer KS, Boop F, Klimo P, DeCupyere M, et al. Patient-derived orthotopic xenografts of pediatric brain tumors: a St. Jude resource. Acta Neuropathol 2020;140:209–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kawauchi D, Robinson G, Uziel T, Gibson P, Rehg J, Gao C, et al. A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer Cell 2012;21:168–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smith SMC, Bianski BM, Orr BA, Harknett G, Onar-Thomas A, Gilbertson RJ, et al. Preclinical modeling of image-guided craniospinal irradiation for very-high-risk medulloblastoma. Int J Radiat Oncol Biol Phys 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rehg J, Ward J. Application of immunohistochemistry in toxicologic pathology of the hematolymphoid system. In: Parker GA, editor. Immunopathology in toxicology and drug development V2. Cham, Switzerland: Humana Press; 2017. p. 489–561. [Google Scholar]
- 22. Vo BT, Kwon JA, Li C, Finkelstein D, Xu B, Orr BA, et al. Mouse medulloblastoma driven by CRISPR activation of cellular Myc. Sci Rep 2018;8:8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Khatamian A, Paull EO, Califano A, Yu J. SJARACNe: a scalable software tool for gene network reverse engineering from big data. Bioinformatics 2019;35:2165–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B, et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 2017;31:737–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hovestadt V, Smith KS, Bihannic L, Filbin MG, Shaw ML, Baumgartner A, et al. Resolving medulloblastoma cellular architecture by single-cell genomics. Nature 2019;572:74–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Infante JR, Cassier PA, Gerecitano JF, Witteveen PO, Chugh R, Ribrag V, et al. A phase I study of the cyclin-dependent kinase 4/6 inhibitor ribociclib (LEE011) in patients with advanced solid tumors and lymphomas. Clin Cancer Res 2016;22:5696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rader J, Russell MR, Hart LS, Nakazawa MS, Belcastro LT, Martinez D, et al. Dual CDK4/CDK6 inhibition induces cell-cycle arrest and senescence in neuroblastoma. Clin Cancer Res 2013;19:6173–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Du X, Wen J, Wang Y, Karmaus PWF, Khatamian A, Tan H, et al. Hippo/Mst signalling couples metabolic state and immune function of CD8alpha(+) dendritic cells. Nature 2018;558:141–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carter RA, Bihannic L, Rosencrance C, Hadley JL, Tong Y, Phoenix TN, et al. A single-cell transcriptional atlas of the developing murine cerebellum. Curr Biol 2018;28:2910–20. [DOI] [PubMed] [Google Scholar]
- 30. Gajjar A, Robinson GW, Smith KS, Lin T, Merchant TE, Chintagumpala M, et al. Outcomes by clinical and molecular features in children with medulloblastoma treated with risk-adapted therapy: results of an international phase III trial (SJMB03). J Clin Oncol 2021;39:822–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kumar R, Smith KS, Deng M, Terhune C, Robinson GW, Orr BA, et al. Clinical outcomes and patient-matched molecular composition of relapsed medulloblastoma. J Clin Oncol 2021;39:807–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hanaford AR, Archer TC, Price A, Kahlert UD, Maciaczyk J, Nikkhah G, et al. DiSCoVERing innovative therapies for rare tumors: combining genetically accurate disease models with in silico analysis to identify novel therapeutic targets. Clin Cancer Res 2016;22:3903–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cook Sangar ML, Genovesi LA, Nakamoto MW, Davis MJ, Knobluagh SE, Ji P, et al. Inhibition of CDK4/6 by palbociclib significantly extends survival in medulloblastoma patient-derived xenograft mouse models. Clin Cancer Res 2017;23:5802–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rusert JM, Juarez EF, Brabetz S, Jensen J, Garancher A, Chau LQ, et al. Functional precision medicine identifies new therapeutic candidates for medulloblastoma. Cancer Res 2020;80:5393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Geoerger B, Bourdeaut F, DuBois SG, Fischer M, Geller JI, Gottardo NG, et al. A phase I study of the CDK4/6 inhibitor ribociclib (LEE011) in pediatric patients with malignant rhabdoid tumors, neuroblastoma, and other solid tumors. Clin Cancer Res 2017;23:2433–41. [DOI] [PubMed] [Google Scholar]
- 36. Geoerger B, Chisholm J, Le Deley MC, Gentet JC, Zwaan CM, Dias N, et al. Phase II study of gemcitabine combined with oxaliplatin in relapsed or refractory paediatric solid malignancies: An innovative therapy for children with cancer european consortium study. Eur J Cancer 2011;47:230–8. [DOI] [PubMed] [Google Scholar]
- 37. Song BS, Seo J, Kim DH, Lim JS, Yoo JY, Lee JA. Gemcitabine and docetaxel for the treatment of children and adolescents with recurrent or refractory osteosarcoma: Korea Cancer Center Hospital experience. Pediatr Blood Cancer 2014;61:1376–81. [DOI] [PubMed] [Google Scholar]
- 38. Álvarez-Fernández M, Malumbres M. Mechanisms of sensitivity and resistance to CDK4/6 inhibition. Cancer Cell 2020;37:514–29. [DOI] [PubMed] [Google Scholar]
- 39. Kruger K, Geist K, Stuhldreier F, Schumacher L, Blumel L, Remke M, et al. Multiple DNA damage-dependent and DNA damage-independent stress responses define the outcome of ATR/Chk1 targeting in medulloblastoma cells. Cancer Lett 2018;430:34–46. [DOI] [PubMed] [Google Scholar]
- 40. Moreira DC, Venkataraman S, Subramanian A, Desisto J, Balakrishnan I, Prince E, et al. Targeting MYC-driven replication stress in medulloblastoma with AZD1775 and gemcitabine. J Neurooncol 2020;147:531–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Forget A, Martignetti L, Puget S, Calzone L, Brabetz S, Picard D, et al. Aberrant ERBB4-SRC signaling as a hallmark of group 4 medulloblastoma revealed by integrative phosphoproteomic profiling. Cancer Cell 2018;34:379–95. [DOI] [PubMed] [Google Scholar]
- 42. Archer TC, Ehrenberger T, Mundt F, Gold MP, Krug K, Mah CK, et al. Proteomics, Post-translational modifications, and integrative analyses reveal molecular heterogeneity within medulloblastoma subgroups. Cancer Cell 2018;34:396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tsesmetzis N, Paulin CBJ, Rudd SG, Herold N. Nucleobase and nucleoside analogues: resistance and re-sensitisation at the level of pharmacokinetics, pharmacodynamics and metabolism. Cancers 2018;10:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang C, Boyson CA, Di Liberto M, Huang X, Hannah J, Dorn DC, et al. CDK4/6 inhibitor PD 0332991 sensitizes acute myeloid leukemia to cytarabine-mediated cytotoxicity. Cancer Res 2015;75:1838–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hurvitz SA, Martin M, Press MF, Chan D, Fernandez-Abad M, Petru E, et al. Potent cell-cycle inhibition and upregulation of immune response with abemaciclib and anastrozole in neoMONARCH, phase II neoadjuvant study in HR(+)/HER2(-) breast cancer. Clin Cancer Res 2020;26:566–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kumarasamy V, Ruiz A, Nambiar R, Witkiewicz AK, Knudsen ES. Chemotherapy impacts on the cellular response to CDK4/6 inhibition: distinct mechanisms of interaction and efficacy in models of pancreatic cancer. Oncogene 2020;39:1831–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ruscetti M, Leibold J, Bott MJ, Fennell M, Kulick A, Salgado NR, et al. NK cell-mediated cytotoxicity contributes to tumor control by a cytostatic drug combination. Science 2018;362:1416–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ruscetti M, M JPt, Mezzadra R, Russell J, Leibold J, Romesser PB, et al. Senescence-induced vascular remodeling creates therapeutic vulnerabilities in pancreas cancer. Cell 2020;181:424–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li J, Jiang J, Wu J, Bao X, Sanai N. Physiologically based pharmacokinetic modeling of central nervous system pharmacokinetics of CDK4/6 inhibitors to guide selection of drug and dosing regimen for brain cancer treatment. Clin Pharmacol Ther 2021;109:494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data for MB002, #2416 and SJMBG3-12-5950 were deposited to the EGA (EGAS00001006001) and the St. Jude Cloud.



![Figure 1. Ribociclib is brain penetrant and decreases pRB, Ki67, and the expression of E2F target genes in human and mouse G3MB tumors. A, Ribociclib plasma and extracellular fluid (ECF) model predictions in the mouse G3MB tumor #2416. B, Pharmacokinetic model structure. Unbound plasma concentrations (open black circles), mean model plasma predictions (solid black line), in tumor ECF concentrations (tECF, open red triangles), and mean model tECF predictions (dashed red line) in the mouse G3MB tumor #2416. CL, CL influx, CL efflux represents the elimination plasma to ECF influx and efflux clearances. OG, oral gavage; CL, clearance. Mice bearing human PDOX MB002 (C–F) or mouse tumor #2416 (G and H) tumors treated with vehicle or ribociclib (100 mg/kg), daily for 5 days and euthanized 4 hours post last dose. C and G, Representative IHC images tumors stained with antibodies against pRbSer807/811 (pRB), Ki67 and activated Caspase-3. D and H, Quantification of the percentage of positive cells for each staining [Mann–Whitney test, n = 7 per group for MB002 (D) and #2416, (H), P ≤ 0.05 (*), P ≤ 0.01 (**)]. Note: two untreated #2416 tumors added to #2416 vehicle group (H). E, Gene Set Enrichment Analysis (GSEA) showed significant depletion of HALLMARK_E2F_TARGETS in ribociclib versus vehicle-treated MB002 tumors. F, Relative mRNA level of selected E2F target genes quantified by qRT-PCR [Mann–Whitney test; n = 4 per group; P ≤ 0.05 (*)].](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/882b/9662921/e70561484f65/1306fig1.jpg)
![Figure 2. Combination therapy of ribociclib with gemcitabine improves survival of mice bearing G3MB tumors. A and B, Treatment schedules for human and mouse models, respectively. C—G, Mice bearing PDOX MB002 tumor cells treated with vehicle (black lines), ribociclib [100 mg/kg, continuous daily by oral gavage (OG), red lines], gemcitabine (60 mg/kg, i.v., day1 then every 2 weeks, blue lines), or the combination (purple lines) until moribund. H–K, Mice bearing mouse tumor #2416 treated with vehicle (black lines), ribociclib [100 or 200 mg/kg, day1–21 (3 weeks), OG, red lines], gemcitabine (60 mg/kg, i.v., day 1 then every 2 weeks, blue lines), or the combination of gemcitabine (60 mg/kg, i.v., day1 then every 2 weeks) with ribociclib (100 or 200 mg/kg, day1–21, OG, purple lines). Mice were treated with sequential cycles of therapy until moribund. C and H, Kaplan–Meier survival plots for all treatment groups. Comparison between treatment groups using log-rank test adjusted for multiple comparisons [not significant (n.s.), adjusted P ≤ 0.05 (*), adjusted P ≤ 0.005 (**), and adjusted P ≤ 0.0005 (***)]. Mice were censored if they died or required humane euthanasia not related to tumor and without preceding neurologic symptoms (e.g., death during sedation for imaging). Mice were imaged twice weekly by bioluminescence imaging (BLI). D and I, BLI pictures at different time points from enrollment through moribund stage represented for one mouse per treatment group. Mice were selected based on tumor and spinal growth median behavior. E, F, and J, BLI increase from enrollment (LOG10 transformation) for brain (E and J) and spine [F; Mann–Whitney test, P ≤ 0.05 (*), P ≤ 0.01 (**)]. For all BLI signal curves, see Supplementary Fig. S3. G and K, Endpoint BLI images of mice with visible spinal signal for MB002 (G), and #2416 (K), based on respective scales. Please note that the endpoint images of the vehicle and ribociclib mice presented in D were the same as in G where they are represented as part of all the mice imaged at endpoint in the study.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/882b/9662921/a733d795ed8e/1306fig2.jpg)