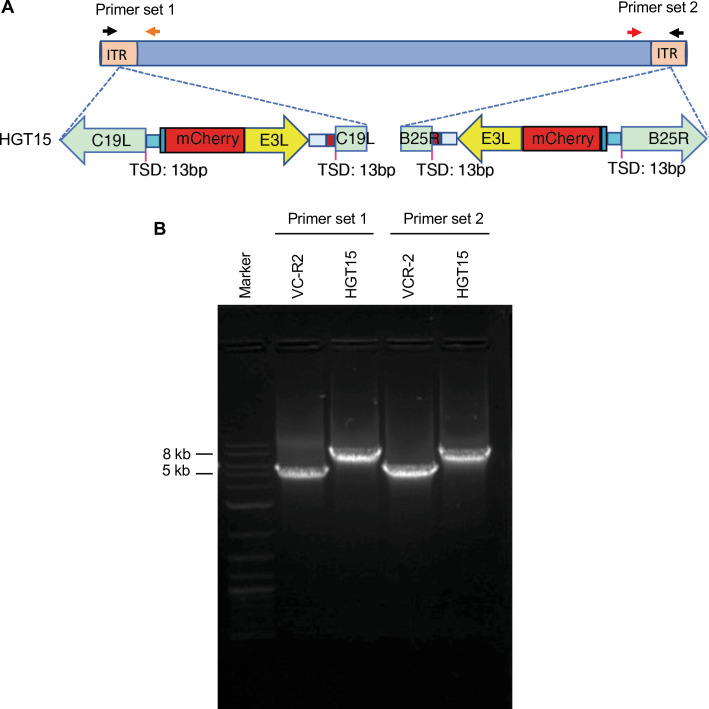
Figure 1. Detection of experimental horizontal gene transfer (HGT) in vaccinia virus.
(A) VACV lacking E3L and K3L cannot replicate in wild-type RK13 cells, but can replicate in cells stably transfected with mCherry-E3L. Virus that acquired E3L can replicate in wild-type RK13 cells (right). (B) Schematic of the mCherry-E3L vector that was stably transfected into RK13 cells. (C) Schematic of the general genetic architecture of horizontally transferred genes identified in VACV isolates. (D) HGT integration sites in the VACV genome. The VACV genome is represented in blue. Genes highlighted above the genome were described to have likely originated from HGT (Hughes and Friedman, 2005; Bratke and McLysaght, 2008). Features shown below the genome are: integration sites into genes (black boxes, blue lines) or into intergenic regions (red lines). The orientation of the transferred genes is indicated by the arrow, colors of arrows indicate the orientation of mCherry-E3L relative to target genes (blue: same direction; red: opposite direction; black arrow: intergenic). (E) Maps of mCherry-E3L integration sites in HGT1–20. Arrows indicate the direction of transcription for VACV and mCherry-E3L. Intergenic regions are depicted in yellow. The position of an integrated U2 small nuclear RNA and the associated poly(A) tract is shown for HGT20 by dashed lines.