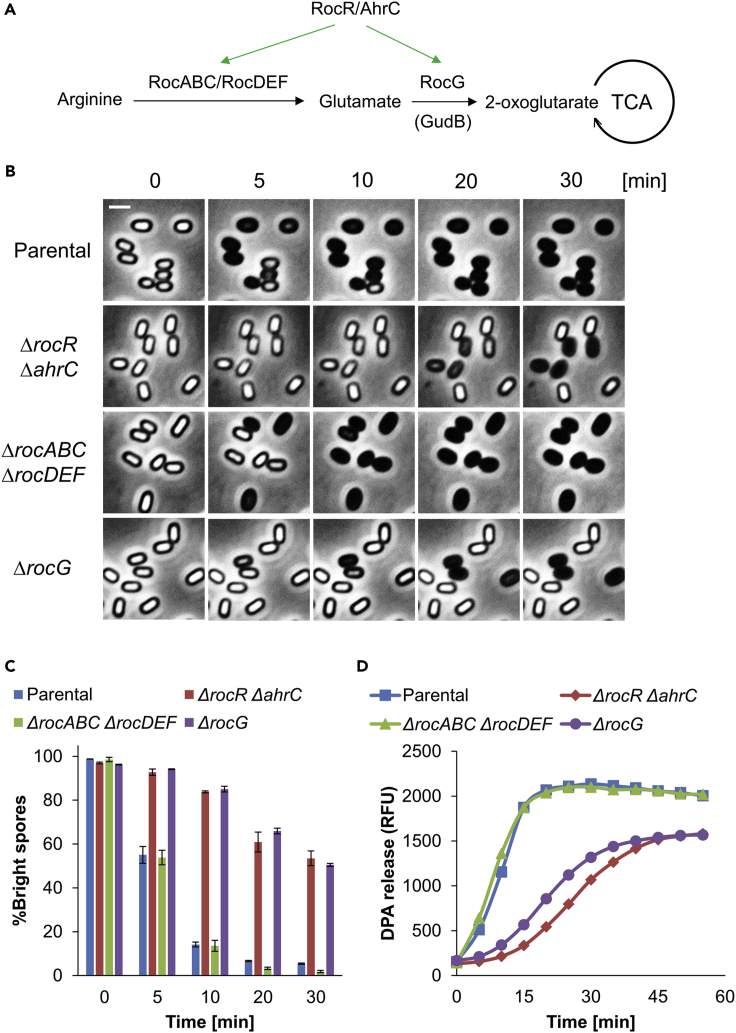
Figure 2.
RocG plays a key role in spore germination
(A) The pathway of RocR-AhrC regulated arginine metabolism in B. subtilis is illustrated. It consists of two stages: (1) Arginine is converted to glutamate by RocABC and RocDEF (RocA-F) enzymes, and (2) RocG dehydrogenase catabolizes glutamate to 2-OG, which participates in the tricarboxylic acid cycle (TCA cycle) (Stannek et al., 2015).
(B) Parental LR33, LR38 (ΔrocR ΔahrC), LR203 (ΔrocABC ΔrocDEF), LR137 (ΔrocG) strains, lacking gudB, were induced to germinate on an agarose pad supplemented with L-Ala (10 mM) (t = 0) and followed by time-lapse microscopy. Shown are phase contrast images captured at the indicated time points from a representative experiment out of three independent biological repeats. Scale bar, 1 μm.
(C) Quantification of the experiment described in (B). Data are presented as percentages of the initial number of the phase bright spores. Shown are average values and SD obtained from three independent biological repeats (n ≥ 300 for each strain).
(D) Spores of LR33 (Parental), LR38 (ΔrocR ΔahrC), LR203 (ΔrocABC ΔrocDEF), LR137 (ΔrocG) strains, lacking gudB, were incubated with L-Ala (10 mM) to trigger germination. DPA release was determined by monitoring the relative fluorescence units (RFU) of Tb3+-DPA. Shown is a representative experiment out of three independent biological repeats.