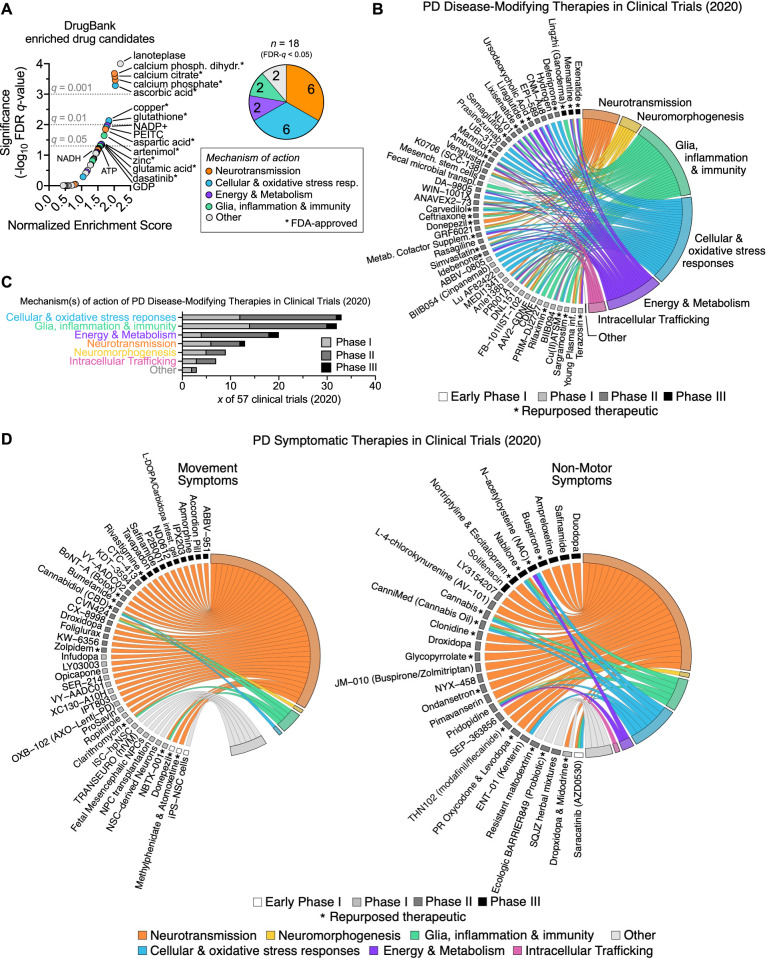
Fig. 6. Prediction of therapeutics for Parkinson’s disease based on dysregulated transcriptomic pathways.
A GSEA against DrugBank’s database of drug-target gene associations90 (n = 5,724 gene sets) identifies 18 therapeutic drugs, each associated with a minimum of 15 target genes, that are significantly (FDR-q < 0.05) overrepresented among PD-dysregulated genes. The top 15 enriched drug candidates with the highest significance and enrichment score are annotated on the right, with FDA-approved drugs indicated by asterisks. The color code indicates each drug’s main mechanism of action in relation to the major biological themes dysregulated in PD reprogrammed neurons (Fig. 5). B, D. Chord plots of disease-modifying (B) and symptomatic (D) PD clinical trial therapies in 2020 (identified by2, left semicircle) and their involvement in dysregulated transcriptomic pathways in PD (right semicircle). Therapies were filtered for redundancy and are grouped by clinical trial phase (see legend). Asterisks indicate repurposed therapeutics. Symptomatic therapies in (D) are categorized into those targeting movement symptoms (left) and those targeting non-motor symptoms (right). C, E. The number of disease-modifying (C) and symptomatic (E) clinical trials targeting the various dysregulated molecular pathways implicated in PD, subcategorized by clinical trial phase.