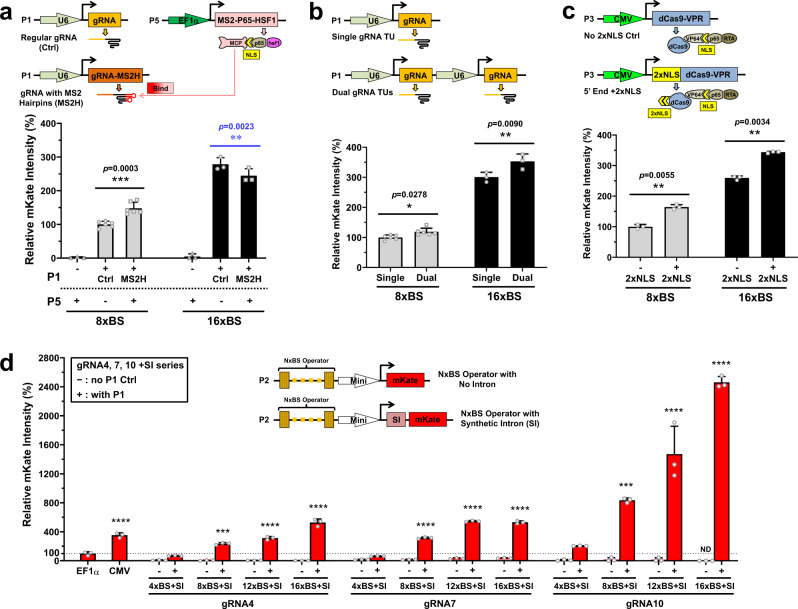
Fig. 3. Genetic control elements added to maximize the expression level of the target gene.
Schematic illustrations depicting individual experiments exhibit only plasmid constructs that were different from P1–P4 in Fig. 1c. Additional genetic control elements were incorporated into the crisprTF promoters (a–d). Experimental groups (+) were transiently transfected with four or five plasmids (P1–P4 or P1–P5), whereas negative controls (−) were transfected without P1. a Synergistic activation mediator (SAM). dCas9-VPR was combined with SAM, another strong synthetic transcriptional activator. Two-tailed paired Student’s t test was performed to compare two P1(+) groups with the same number of gRNA BS (with the 8× BS operator, MS2H gRNA vs. control gRNA, p = 0.0003; with the 16× BS operator, MS2H gRNA vs. control gRNA, p = 0.0023) b Dual gRNA transcriptional units (TUs). An extra gRNA10 TU was added to increase gRNA expression (two-tailed paired Student’s t test; with the 8× BS operator, dual gRNA vs. single gRNA, p = 0.0278; with the 16× BS operator, dual gRNA vs. single gRNA, p = 0.0090). c An additional 2× nuclear localizing sequence (NLS) in the crisprTF was added at the 5′-end of dCas9-VPR (two-tailed paired Student’s t test; with the 8× BS operator, 2× NLS vs. control, p = 0.0055; with the 16× BS operator, 2× NLS vs. control, p = 0.0034). d A synthetic intron (SI) was added at the 5′ UTR of the target gene. The SI was incorporated into four synthetic operators of the gRNA4, 7, and 10 series (4×, 8×, 12×, and 16× BS), respectively. Red solid bars represent experimental groups (+); paired red hollow bars represent corresponding control groups without gRNA (−) (one-way ANOVA with multiple comparisons corrected by Dunnett test for each gRNA series with the same EF1α and CMV control groups; CMV, gRNA4 P1(+) 8×, 12×, and 16× BS with SI, gRNA7 P1(+) 8×, 12×, and 16× BS with SI, and gRNA10 P1(+) 8×, 12×, and 16× BS with SI vs. EF1α: p < 0.0001, p = 0.0003, p < 0.0001, p < 0.0001, p < 0.0001, p < 0.0001, p < 0.0001, p = 0.0006, p < 0.0001, and p < 0.0001, respectively). All data represent the mean ± SD (n ≥ 3). For increased expression (marked in black): *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; for decreased expression (marked in blue): **p < 0.01; ND not detected. Source data are provided as Source Data files.