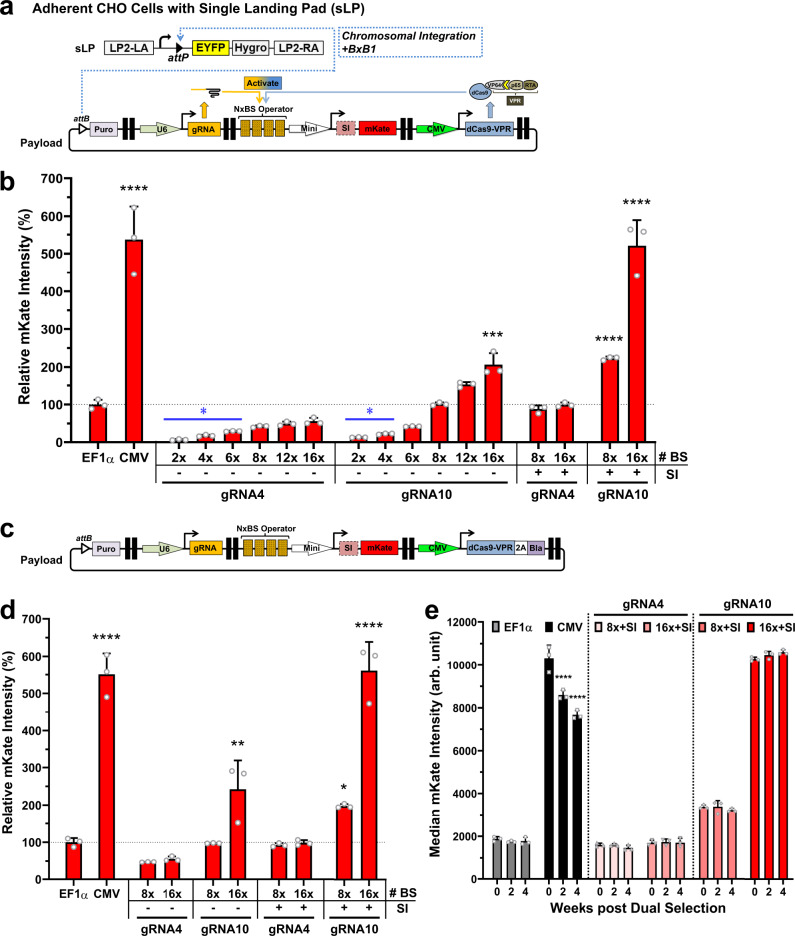
Fig. 4. Genomic integration and long-term precision gene expression in CHO landing pad cells.
a A schematic illustration of an integration gene circuit and BxB1 recombinase-mediated, site-specific integration in an engineered, adherent CHO cell line with a single landing pad (sLP). Positive integration control circuits had a central TU with an EF1α or CMV promoter driving mKate expression and two flanking dummy TUs with no gene expression in the same architecture. b The mKate signal intensities of the chromosomally integrated crisprTF promoter circuits in sLP-CHO cells relative to the integrated EF1α control circuit (one-way ANOVA with multiple comparisons corrected by Dunnett test; CMV, gRNA4 2×, 4×, and 6× BS without SI, and gRNA10 2×, 4×, and 16× BS without SI vs. EF1α: p < 0.0001, p = 0.0031, p = 0.0111, p = 0.0414, p = 0.0066, p = 0.0191, and p = 0.0006, respectively; gRNA10 8× and 16× BS with SI vs. EF1α: p < 0.0001 and p < 0.0001, respectively). c A schematic illustration of an integration circuit with the addition of a 3′ flanking selection marker (blasticidin) into dCas9-VPR, linked by a 2A self-cleavage peptide, to increase stability of target gene expression in long-term culture. A number of integration circuits from the gRNA4 and gRNA10 series were selected for the modification. d The mKate signal intensities of the chromosomally integrated crisprTF promoter circuits after dual selection with puromycin and blasticidin (one-way ANOVA with multiple comparisons corrected by Dunnett test; CMV and gRNA10 16× BS without SI vs. EF1α: p < 0.0001 and p = 0.0020, respectively; gRNA10 8× and 16× BS with SI vs. EF1α: p = 0.0446 and p < 0.0001, respectively). e The mKate expression levels with two of the strongest circuits from each gRNA series over the course of 4 weeks following dual selection. All four circuits examined displayed no noteworthy change in mKate signals at 2 or 4 weeks (all p > 0.05), similar to the EF1α control (two-way ANOVA with multiple comparisons corrected by Dunnett test; CMV: 0 vs. 2 weeks: p < 0.0001, 0 vs. 4 weeks: p < 0.0001). All data represent the mean ± SD (n = 3) (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). Source data are provided as Source Data files.