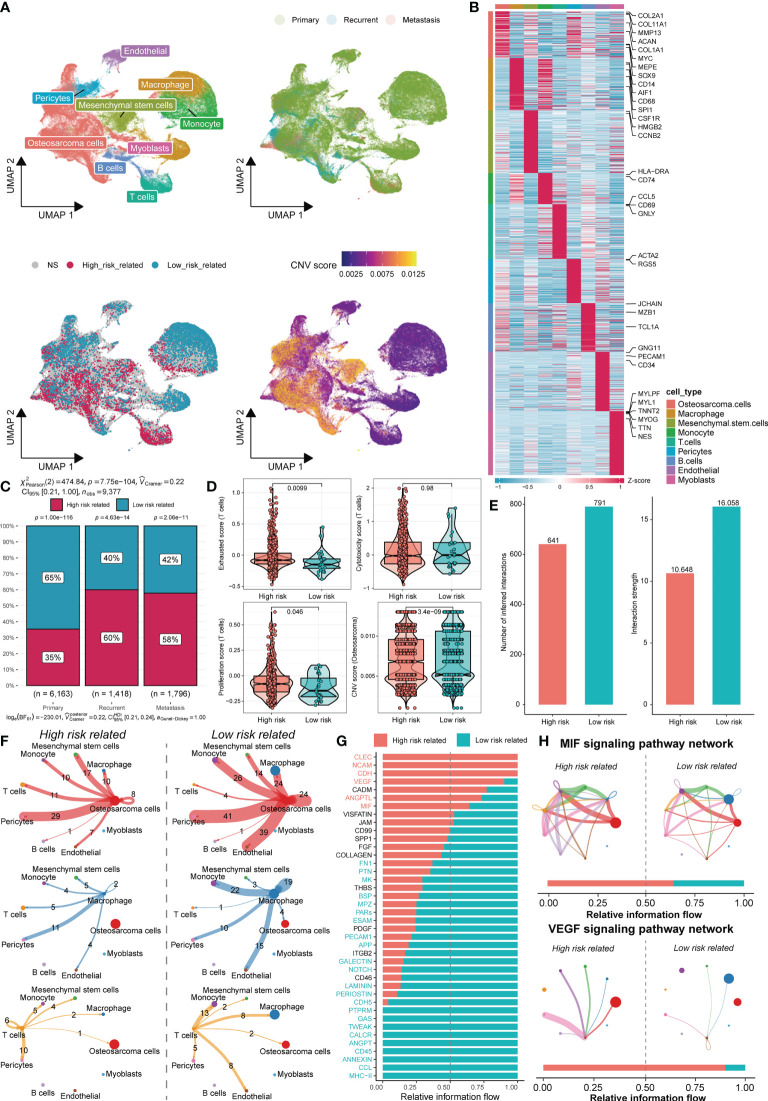
Figure 6.
TME differences in cells associated with different risk scores. (A) UMAP analysis identified nine major cell types in osteosarcoma samples (top left), sample groupings involving primary, recurrent, and metastasis (top right), associated cells by SCISSOR risk groupings (bottom left), and CNV score inferred by the infercnv-related heatmap (bottom right). (B) The DEGs and marker genes for nine major cell types are shown. The colors in the top and side bars indicate specific cell clusters. (C) A bar graph depicts the proportion of cells in different sample groupings that are associated with the risk score. (D) Boxplots depict the exhaustion (top left), toxicity (top right), and proliferation (bottom left) scores of T cells in the two risk score groups. The difference in the CNV score between osteosarcoma cells is shown (bottom right). (E) Differences in the number of cells associated with different risk scores and the intensity of their cellular communication. (F) Circos plots depict putative ligand–receptor interactions between T cells from the high-risk (left) and low-risk (right) associated cell groups and other cell clusters. Branches connect pairs of interacting cell types and indicate the number of events in the graph. (G) In the inferred network, the overall information flow differences for the different risk-group-associated cells are shown. (H) Circos plots show the MIF signaling pathway and VEGF signaling pathway of related cells in different risk groups.