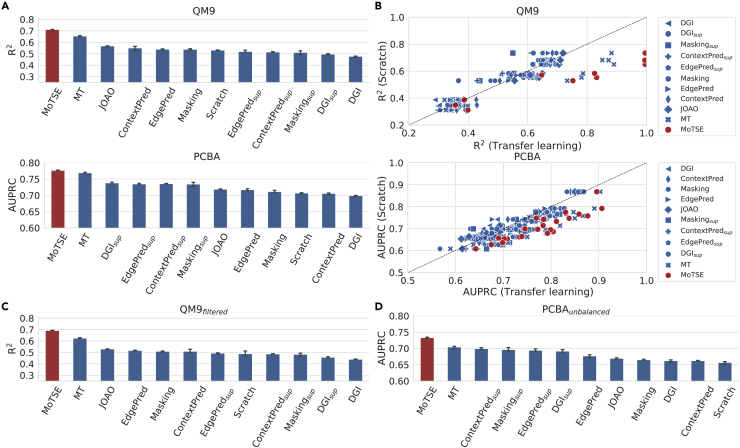
Figure 3.
MoTSE outperforms baseline methods and alleviates negative transfer on the QM9 and PCBA datasets
(A) The prediction performance of MoTSE and eleven baseline methods on the QM9 and PCBA datasets, measured in terms of R2 and AUPRC, respectively.
(B) The prediction performance of eleven transfer learning methods versus that of the Scratch method on the QM9 and PCBA datasets.
(C) The comparison results of R2 between MoTSE and eleven baseline methods on the QM9 dataset after filtering every molecule from the test set if it has a Tanimoto similarity score greater than 0.8 to any molecule in the training set (also see Figure S4A).
(D) The comparison results of AUPRC between MoTSE and eleven baseline methods on an unbalanced PCBA dataset with only positive samples (also see Figure S4B).