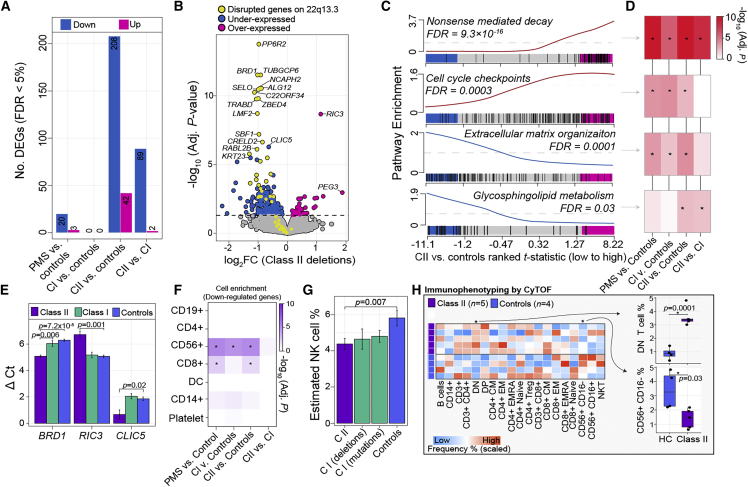
Figure 2.
Altered peripheral blood gene expression profiles in class II mutations
(A) The total number of differentially expressed genes (DEGs) (y axis) for each comparison (x axis). Each analysis adjusted for sex and age as covariates.
(B) Volcano plot of class II DEGs relative to unaffected controls depicting log2 fold change (log2FC; x axis) and –log10 FDR adjusted p value (y axis). The horizontal line indicates FDR < 5%. Genes in yellow are the 52 genes expressed in blood that are affected by large class II mutations in PMS. Genes in blue are all other downregulated genes and those in pink are all other upregulated genes.
(C) Four representative pathway enrichment scores (y axis) of class II DEGs according to ranked t statistics, high (pink) to low (blue) (x axis). All enrichment results can be found in Table S2.
(D) The resulting FDR adjusted p value enrichment for all differential comparisons reveals shared and unique gene set enrichment among class II and class I mutations.
(E) qRT-PCR validation of three target genes across four technical replicates (used to generate standard error bars) per group: BRD1 (a downregulated gene on chr 22); RIC3 (an upregulated gene on chr 11); and CLIC5 (a downregulated gene on chr 6). A Student’s t test was used to test delta CT values significant differences.
(F) CAMERA cell-type enrichment of underexpressed DEGs (x axis) according to seven immune cell types (y axis) reveals strong enrichment of CD56+ genes.
(G) CIBERSORTx cell-type predictions reveal a significant reduction in the frequency CD56+ cells among class II mutations. Standard error bars summarize variability across each respective group.
(H) CyTOF validates estimated cell-type proportions on a subset of controls and participants with class II mutations. Scaled frequencies across all participants for major and minor immune cell populations are presented in heatmap form (right). Boxplots of the two immune populations with significant differences (p < 0.05, linear model) associated with class II mutations (left).