Figure 2.
Mito-Cas9 system mediated efficient knockin of exogenous ssODNs into mtDNA
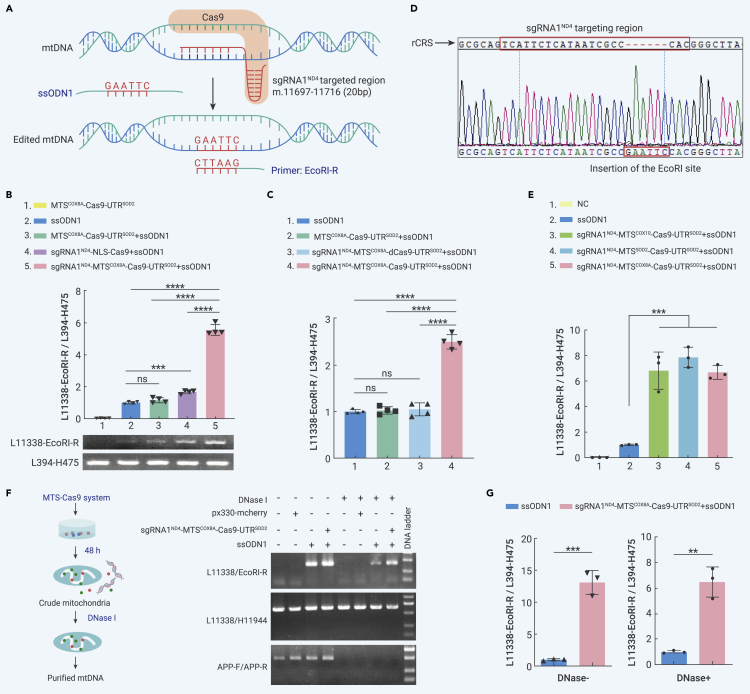
(A) Design of mito-Cas9-mediated knockin system.
(B) Quantification of knockin efficiency of mito-Cas9. (Upper) Results of qRT-PCR. HEK293T cells were transfected with or without a combination of Cas9 constructs and ssODN1. Content of mtDNA with successful knockin of EcoRI site (edited mtDNA, amplified by primer pair L11338/EcoRI-R (L11338-EcoRI-R)) was normalized to whole mtDNA (total mtDNA, amplified by primer pair L394/H475 (L394-H475)), using total genomic DNA as the template. (Below) Agarose gel image showing representative PCR product for each group of cells with different transfections.
(C) Knockin efficiency of mito-Cas9 constructs with catalytically dead Cas9 (dCas9): sgRNA1ND4-MTSCOX8A-dCas9-UTRSOD2, expressing SpCas9 protein with mutations p.D10A and p.H840A. Knockin efficiency was quantified using the same qRT-PCR procedure as in (B), with total genomic DNA from respective transfected cells as the template.
(D) Sequencing chromatogram of EcoRI site-specific PCR product. Targeting region of sgRNA1ND4 is marked with a box on the mtDNA revised Cambridge reference sequence (rCRS).
(E) Knockin efficiency of different mito-Cas9 constructs with different MTSs. HEK293T cells were transfected with ssODN1 and mito-Cas9 construct fused with indicated MTSs of mitochondrial genes COX8A, COX10, or SOD2. Knockin efficiency was quantified using the same qRT-PCR procedure as in (B), with total genomic DNA from respective transfected cells as the template. NC, cells without transfection.
(F) DNase protection assay confirmed the mitochondrial source of EcoRI site-specific PCR product amplified from edited mtDNA in mitochondria. Crude mitochondria were extracted from HEK293T cells transfected with sgRNA1ND4-MTSCOX8A-Cas9-UTRSOD2 and ssODN1 for 48 h, then treated with DNase I at 37°C for 1 h. PCR amplifications for nuclear APP (amplified by primer pair APP-F/APP-R), total mtDNA (amplified by primer pair L11338/H11944), and edited mtDNA (amplified by primer pair L11338/EcoRI-R) were performed using mtDNA template extracted from mitochondria with or without DNase I treatment, respectively.
(G) Quantification of knockin efficiency in mtDNAs from crude mitochondria treated with DNase I (DNase+) or without DNase I (DNase−). Bars are mean ± SD. ns, not significant; ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; one-way ANOVA test adjusted by Tukey’s multiple comparisons test for (B, C, and E); two-tailed Student's t test for (G).