Figure 6. Tcf1 and cooperate with CTCF to regulate chromatin interactions in CD8+ T cells.
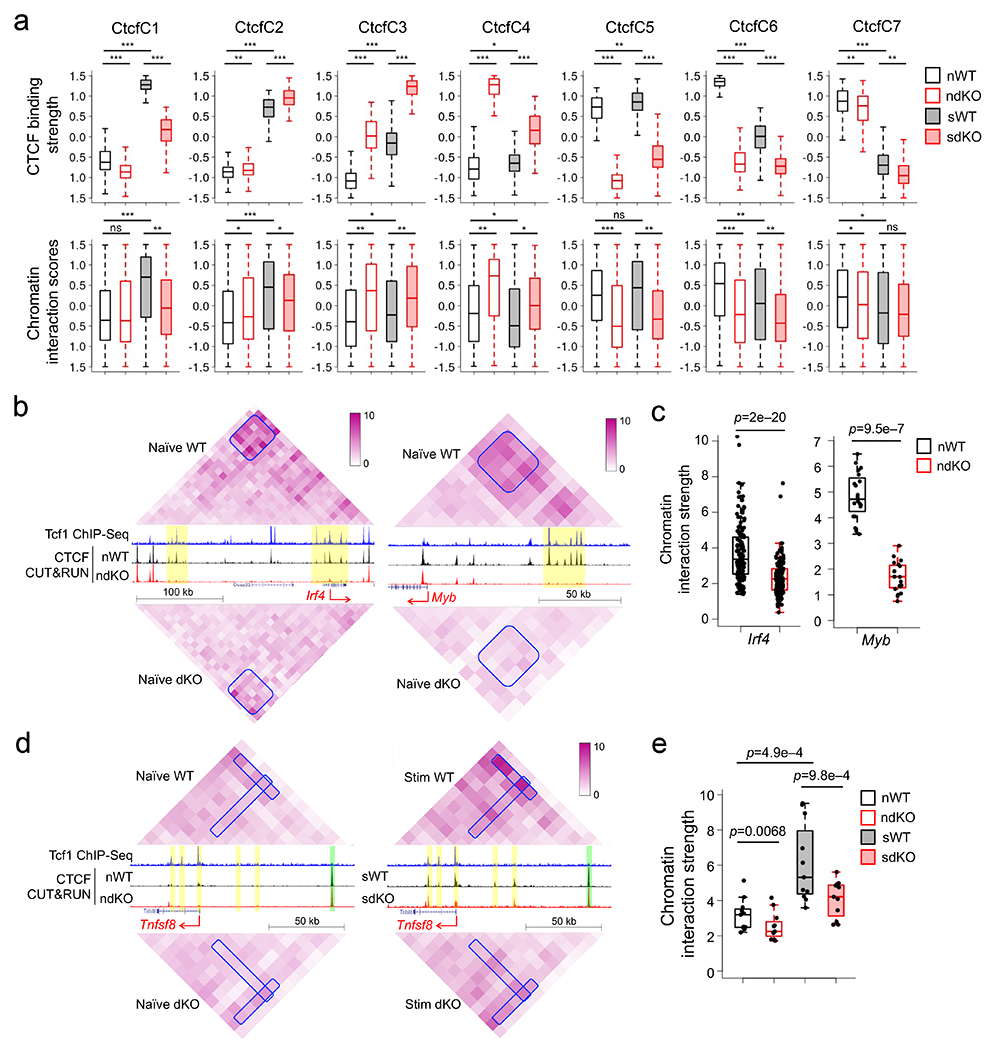
a. Box plots summarizing CTCF binding strength in each CTCF cluster as defined in Fig. 2f (top) and chromatin interaction scores of anchors harboring corresponding CTCF sites in each cluster (bottom), with both parameters z-score-transformed and plotted for naïve WT and dKO (nWT and ndKO, respectively), and IL-7+IL-15-stimulated WT and dKO (sWT and sdKO, respectively) CD8+ T cells. *, p<0.05; **, p<1e–10; ***, p<1e–30 by one-sided paired Wilcoxon test. b. Diamond graphs showing distance-normalized chromatin interactions in naïve WT (top) and dKO CD8+ T cells (bottom) at the Irf4 (left) and Myb (right) gene loci, as displayed on WashU epigenome browser, with blue boxes denoting chromatin interaction ‘patches’ showing marked changes. Shown in the middle are gene structures, Tcf1 ChIP-seq tracks in naïve WT, CTCF CUT&RUN tracks in naïve WT and dKO CD8+ T cells, with yellow bars denoting Tcf1+Lef1-dependent CTCF binding sites. c. Box plots summarizing the chromatin interaction strength for each pair of 10-kb anchors within the interaction ‘patches’ at the Irf4 or Myb gene locus in naïve WT and dKO CD8+ T cells, with p-value determined using one-sided paired Wilcoxon test. d. Diamond graphs showing distance-normalized chromatin interactions in naïve (left) and IL-7+IL-15-stimulated (right) WT (top) and dKO CD8+ T cells (bottom) at the Tnfsf8 gene locus, with blue boxes denoting chromatin interaction ‘stripes’ with marked changes. Shown in the middle are gene structure, Tcf1 ChIP-seq tracks in naïve WT, CTCF CUT&RUN tracks in naïve (left) and IL-7+IL-15-stimulated (right) WT and dKO CD8+ T cells, with yellow and green bars denoting dynamic and constitutive CTCF binding sites, respectively. e. Box plots summarizing the chromatin interaction strength within the interaction ‘stripes’ at the Tnfsf8 gene locus in naïve or IL-7+IL-15-stimulated WT and dKO CD8+ T cells, with p-value determined using one-sided paired Wilcoxon test. Parameters for box plots (a, c, e) are the same as Fig. 3b. Color scale (b, d) denotes chromatin interaction strength.
