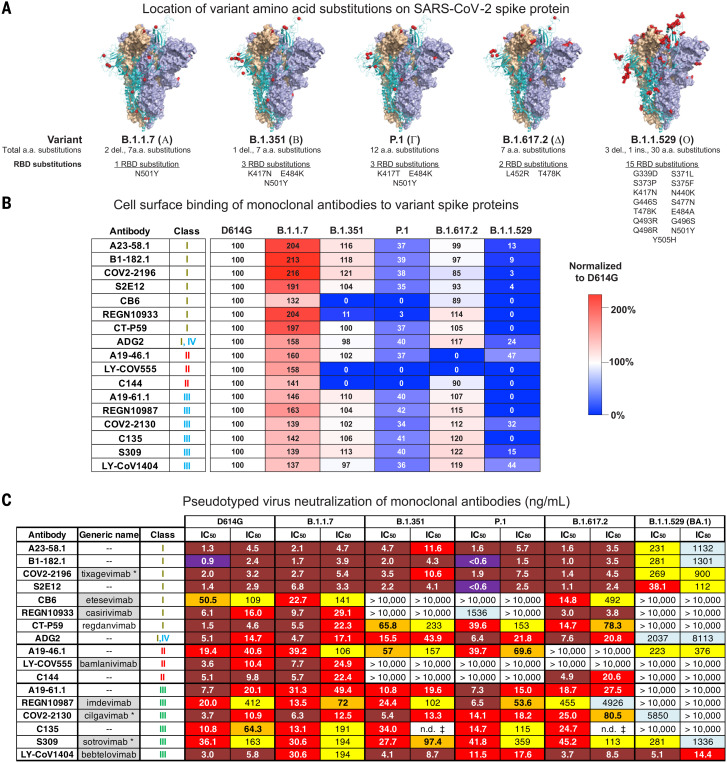
Fig. 2. SARS-CoV-2 monoclonal antibody binding and neutralization.
(A) Models of SARS-CoV-2 WA-1 spike protein (PDB: 6XM3) with the locations of substitutions present in variants indicated as red dots. Also indicated is the total number of amino acid substitutions and the number and locations of RBD substitutions in VOC spike proteins. (B) Full-length spike proteins from the indicated SARS-CoV-2 variants were expressed on the surface of transiently transfected 293T cells, and binding to indicated monoclonal antibodies was assessed by means of flow cytometry. Antibody mean fluorescence intensity (MFI) binding signal was adjusted according to spike protein expression level (fig. S4). Shown is the ratio of the adjusted antibody MFI binding to the indicated spike expressing cells to the adjusted MFI of the same antibody bound to D614G spike–expressing cells. The data are expressed as a percentage. Shown is a representative experiment (n = 2 replicates). (C) Lentiviruses pseudotyped with SARS-CoV-2 spike proteins from D614G, B.1.1.7, B.1.351, P.1, B.1.617.2, or B.1.1.529 (BA.1) were incubated with serial dilutions of the indicated antibodies, and IC50 and IC80 values were determined. S309 was tested on 293 flpin-TMPRSS2-ACE2 cells, whereas all the other antibodies were tested on 293T-ACE2 cells. Ranges are indicated with white (>10,000 ng/ml), light blue (>1000 to ≤10,000 ng/ml), yellow (>100 to ≤1000 ng/ml), orange (>50 to ≤100 ng/ml), red (>10 to ≤50 ng/ml), maroon (>1 to ≤10 ng/ml), and purple (≤1 ng/ml). n.d. ‡, not determined because of incomplete neutralization that plateaued at <80% (fig. S5B). Where available, generic names of antibodies under therapeutic investigation are shown. Gray shading indicates antibodies that previously or currently have received Emergency Use Authorization from the US Food and Drug Administration. Generic names with an asterisk indicate therapeutic antibody products with the same binding regions as those of the antibodies being tested but containing amino acid changes in their Fc domains.