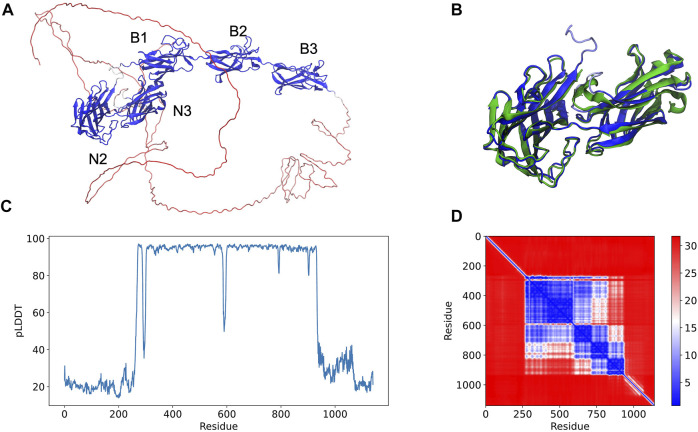
FIGURE 1.
Full-length structure prediction of S. aureus serine-aspartate repeat protein (SdrE, Uniprot ID: Q932F7). (A) Top ranked SdrE model is represented in cartoon and its different domains are indicated. The protein is colored by the pLDDT scores generated by AlphaFold 2 where dark blue represent regions with very high quality (pLDDT > 90) and red represent regions with very low quality (pLDDT < 50). (B) Structural alignment between the N2 and N3 regions of the AlphaFold 2 model (dark blue) and SdrE crystallographic structure (cyan, PDB ID: 5WTA). (C) By residue pLDDT scores for the generated SdrE models. (D) Predicted alignment error (PAE) for the best ranked model. The color at (x, y) corresponds to the expected distance error in residue x’s position, when the prediction and true structure are aligned on residue y.