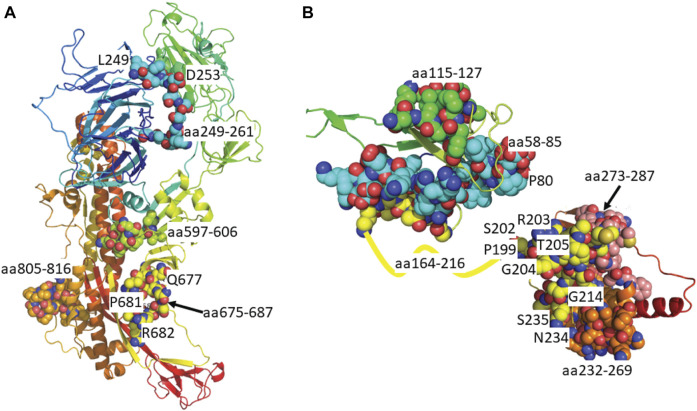
FIGURE 3.
Predicted epitopes mapped to protein structure of Surface Glycoprotein and Nucleoprotein. (A) X-ray crystal structure of the Surface Glycoprotein (Cai et al., 2020) coloured via spectrum (N to C terminal) with the structured B cell epitopes highlighted via spheres; aa294-261 (cyan), aa597-606 (green), aa675-687 (yellow) and aa805-816 (orange). Also labelled are the mutation sites; L249, D253, Q677, P681 and R682. (B) Structure of the nucleoprotein coloured via spectrum (N to C terminal). The N-terminal regions are from the X-ray crystal structure (PDB code: 6vyo) followed by an unstructured linear (yellow line) to a C-terminal homology model. The structural B cell epitopes are highlighted via spheres; aa58-85 (cyan), aa115-127 (green), aa164-216 (yellow), aa232-269 (orange) and aa273-287 (salmon). Also labelled is the location of known mutation sites; P80, P199, R201, S202, R203, T205, G214, N234 and S235.