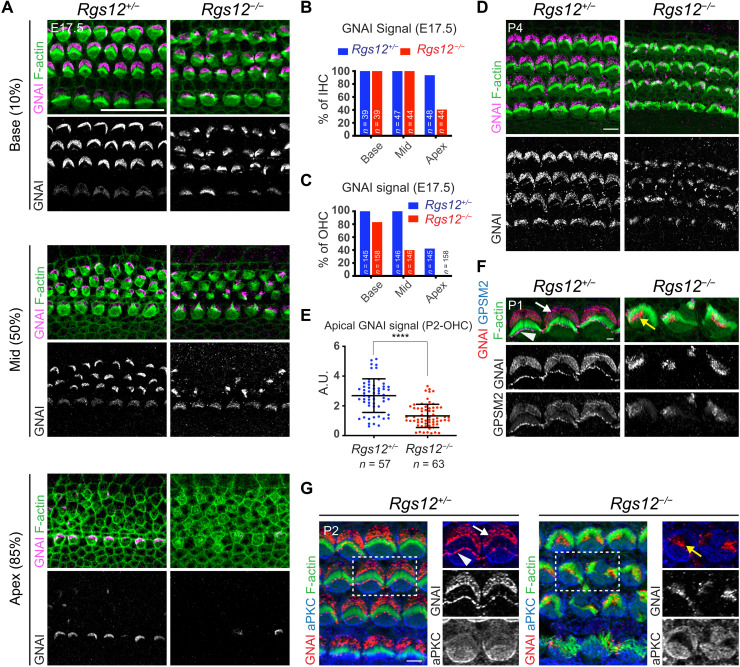
Fig. 3. Disruption of the apical blueprint proteins GPSM2-GNAI and aPKC in Rgs12 mutant HCs.
(A) GNAI immunolabeling at E17.5 at different cochlear positions (% of distance from the cochlear base). In Rgs12 mutants, GNAI is missing (apex, 85% position), delayed (mid, 50%), and fails to encompass the whole bare zone (base, 10%) in progressively more mature HCs. (B and C) Proportion of E17.5 IHCs (B) and OHCs (C) where an apical GNAI signal is detected by cochlear position. n = HCs, N = 3 animals. (D) GNAI immunolabeling. (E) Total GNAI signals at the apical HC surface. N = 3 animals. Means ± SD. t test, ****P < 0.0001. (F) GNAI and GPSM2 coimmunolabeling in OHC2. GNAI and GPSM2 at the bare zone (arrow) and stereocilia tips (arrowhead) are absent in Rgs12 mutants and instead match subregions of the hair bundle (yellow arrow). (G) GNAI and aPKC coimmunolabeling. Boxed OHCs are magnified on the right. In Rgs12 mutants, aPKC extends laterally to encompass the entire flat HC surface, and GNAI is found at the position of the aPKC-negative hair bundle (yellow arrow). Unlike indicated differently, in all figures, arrows indicate protein enrichment at the bare zone and arrowheads indicate enrichment at stereocilia tips. A.U., arbitrary units. Scale bars, 20 μm (A), 5 μm (D and G), and 0.5 μm (F).