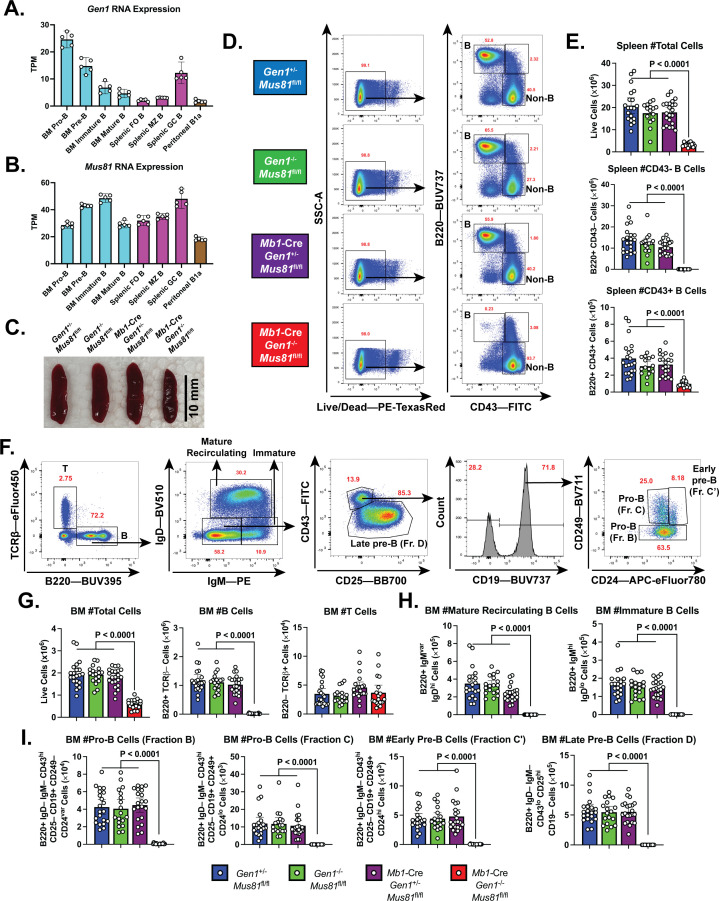
Figure 1. B cell development in the bone marrow (BM) and spleen of Mb1-Cre Gen1−/− Mus81fl/fl mice.
(A, B) mRNA expression of Gen1 (A) and Mus81 (B) in developing and mature B cell subsets in the BM, spleen, and peritoneal cavity. FO: follicular, MZ: marginal zone (GEO Accession: GSE72018). (C) Spleens harvested from 4-month-old mice of the indicated genotypes. (D, E) Gating strategy (D) and absolute quantification (E) of live splenocytes, splenic B220+CD43–B, and B220+CD43+B cells. (F) Gating strategy of total B (B220+TCRβ–), total T (B220–TCRβ+), mature recirculating, immature, pro-B (fractions B and C), early pre-B (fraction C′), and late pre-B cells (fraction D). (G–I) Absolute quantification of BM cellularity, total B, and total T cell populations (G), of mature recirculating and immature B cell populations (H), and of B cell populations belonging to fractions B to D (I). Data in (E) and (G–I) are from four independent experiments with 18–22 mice per genotype. Bars display the arithmetic mean and error bars represent the 95% confidence interval of the measured parameters. P values were enumerated using ordinary one-way ANOVA analysis with Dunnett’s multiple comparisons test without pairing wherein all means were compared to the Mb1-Cre Gen1−/− Mus81fl/fl group. TPM, transcripts per million.