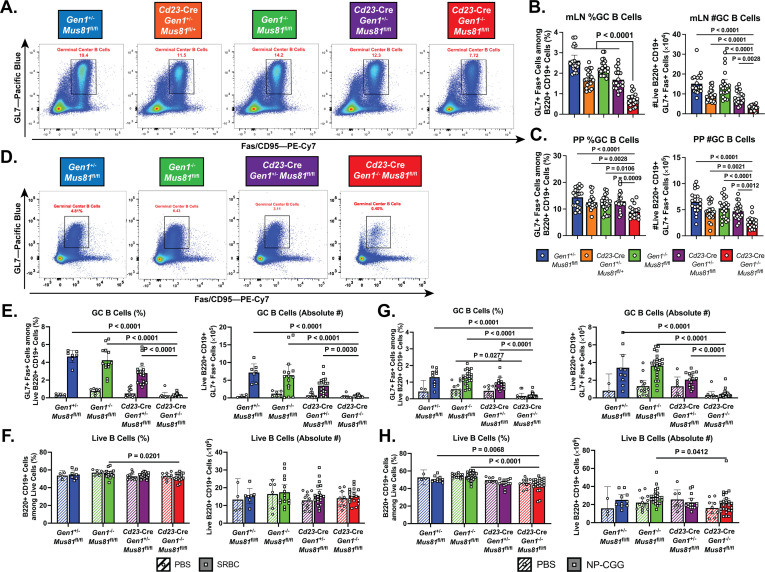
Figure 2. Homeostatic and induced GC responses in Cd23-Cre Gen1−/− Mus81fl/fl mice.
(A–C) Homeostatic GC response in mesenteric lymph nodes and Peyer’s patches. (A) Flow cytometric plots depicting the GL7+Fas+GC B cells in the Peyer’s patches of mice for each indicated genotype. (B, C) Quantification of the frequencies and absolute numbers of the GL7+Fas+GC B cell population in the mesenteric lymph nodes (B) and Peyer’s patches (C). (D–F) Evaluation of GC response during SRBC challenge. (D) Representative plots of the GL7+Fas+GC B cell population isolated from the spleen of mice immunized with SRBC for each indicated genotype. (E, F) Quantification of the frequencies and absolute numbers of GC B cells (E) and of total B cells (F) in the spleen of SRBC-immunized mice. (G, H) Assessment of induced GC response upon NP-CGG challenge in the Cd23-Cre Gen1−/− Mus81fl/fl mice at day 21 post-immunization. (G) Quantification of the percentage and absolute count of GL7+Fas+GC B cells in the spleen. (H) Frequencies and absolute numbers of live B220+ B cells in the spleen of PBS-treated and NP-CGG-immunized mice. Data in (B) and (C) are from four independent experiments with 11–21 mice per genotype. Data in (E) and (F) are from three independent experiments with 4–9 mice per genotype for the PBS group and 7–20 mice per genotype for the SRBC group. Data in (G) and (H) are from three independent experiments with 5–13 mice per genotype in the PBS group and 10–25 mice in the NP-CGG group. Bars represent the arithmetic mean and the error bars depict the 95% confidence interval of the measured parameters. For (B) and (C), p values were computed by ordinary one-way ANOVA analysis with Dunnett’s multiple comparisons test without pairing in which the means were compared to the Cd23-Cre Gen1−/− Mus81fl/fl group. For (E–H), ordinary two-way ANOVA analysis with Dunnett’s multiple comparisons test without pairing was used to calculate the p values. All means were compared within each treatment group to the Cd23-Cre Gen1−/− Mus81fl/fl cohort.