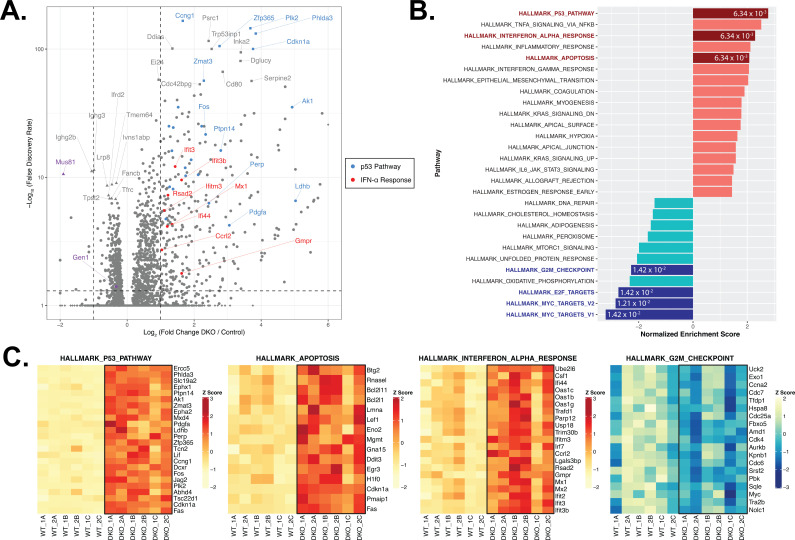
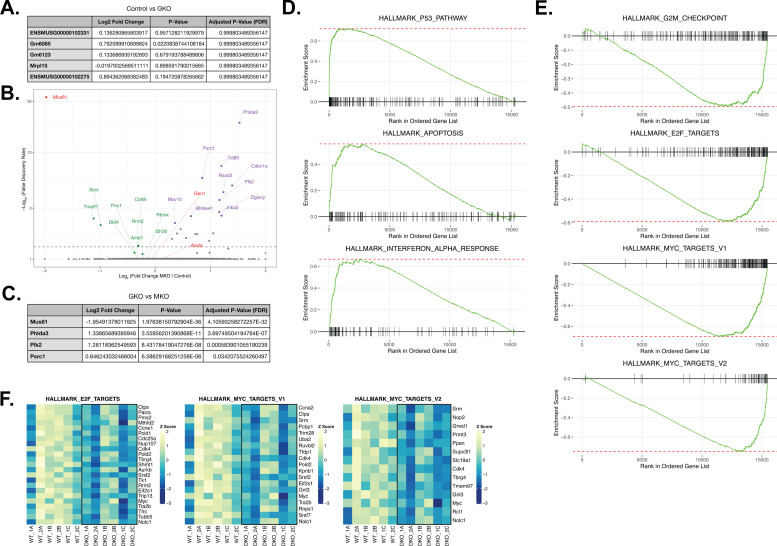
Figure 4. RNA-seq and gene set enrichment (GSEA) analyses of activated control and DKO B cells.
(A) Volcano plot depicting the differential gene expression between control and DKO cells. Labeled squares indicate the top 15 most significantly upregulated genes and the labeled triangles are the 10 most downregulated genes in DKO cells. Using the Hallmark Gene Sets as reference, genes labeled in blue are categorized as genes in the p53 pathway while those labeled in red are defined as interferon alpha (IFN-α) response genes. Purple symbols mark Gen1 and Mus81. (B) Graph depicting the list of Hallmark Gene Sets that are differentially expressed between control and DKO cells (FDR<0.05). Value in each bar denotes the FDR for that gene set. (C) Heatmaps displaying the relative expression of genes within the indicated Hallmark Gene Sets that meet the expression cutoff (log2 fold change>1 for p53 pathway; >0.5 for apoptosis and interferon alpha response; <–0.3 for G2/M checkpoint; with FDR<0.05). Data are from six mice per genotype. The labels ‘1’ and ‘2’ represent male and female mice, respectively, and ‘A’ to ‘C’ indicate the three experimental ex vivo groups.