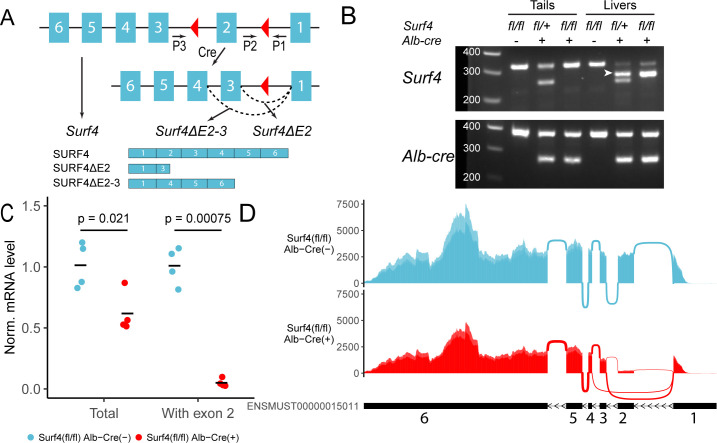
Figure 1. Generation of hepatocyte-specific Surf4 deficient mice.
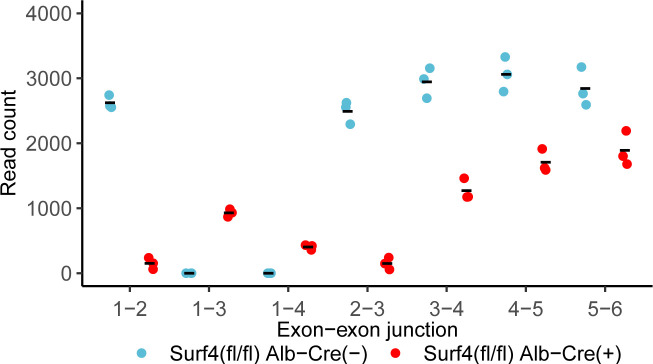
(A) Schematic presentation of the Surf4 conditional allele. Blue rectangles represent exons and black line segments represent introns. Red triangles denote loxP sites. Expression of a Cre recombinase leads to excision of exon 2, which results in the generation of a Surf4 mRNA lacking exon 2 (Surf4ΔE2) or both exon 2 and 3 (Surf4ΔE2-3). Surf4ΔE2 mRNA is translated into a truncated SURF4 that is only 22 amino acids in length. Surf4ΔE2-3 mRNA restores the reading frame, producing an internally truncated protein missing the 88 amino acids encoded by exon 2 and 3. P1, P2, P3 indicate the positions for Surf4 genotyping primers (Supplementary file 1). Dashed arcs represent splicing events. Exons and introns are not drawn to scale. (B) Agarose gel electrophoresis of PCR products generated using genomic DNA (gDNA) isolated from mouse tails and livers and primers P1-3 shown in (A) (Figure 1—source data 1). For Surf4 genotyping, the wild type allele produces a smaller PCR product whereas the conditional allele produces a larger amplicon. Excision of exon 2 results in the generation of a PCR product of intermediate size (white arrowhead) that is present in gDNA isolated from the livers of Alb-Cre+ mice only. For Alb-Cre genotyping, presence of the Cre transgene results in a smaller PCR product. The upper band represents the amplification of an internal control (Supplementary file 1). (C) Quantification of normalized (Norm.) Surf4 mRNA abundance by quantitative PCR of liver cDNA from control (Surf4fl/fl Alb-Cre-) and Surf4fl/fl Alb-Cre+ mice (n=4 per genotype). Crossbars represent the mean normalized abundance in each group. The denoted p-values were calculated by two-sided Student’s t-test. (D) Density plots of RNA-seq reads mapping along exons and exon-exon junctions of Surf4 mRNA. Surf4fl/fl Alb-Cre+ mice have lower overall read counts due to incomplete nonsense mediated mRNA decay. Arcs between exons represent splicing events and line thickness is proportional to read count. Exact read count for each junction is presented in Figure 1—figure supplement 1.