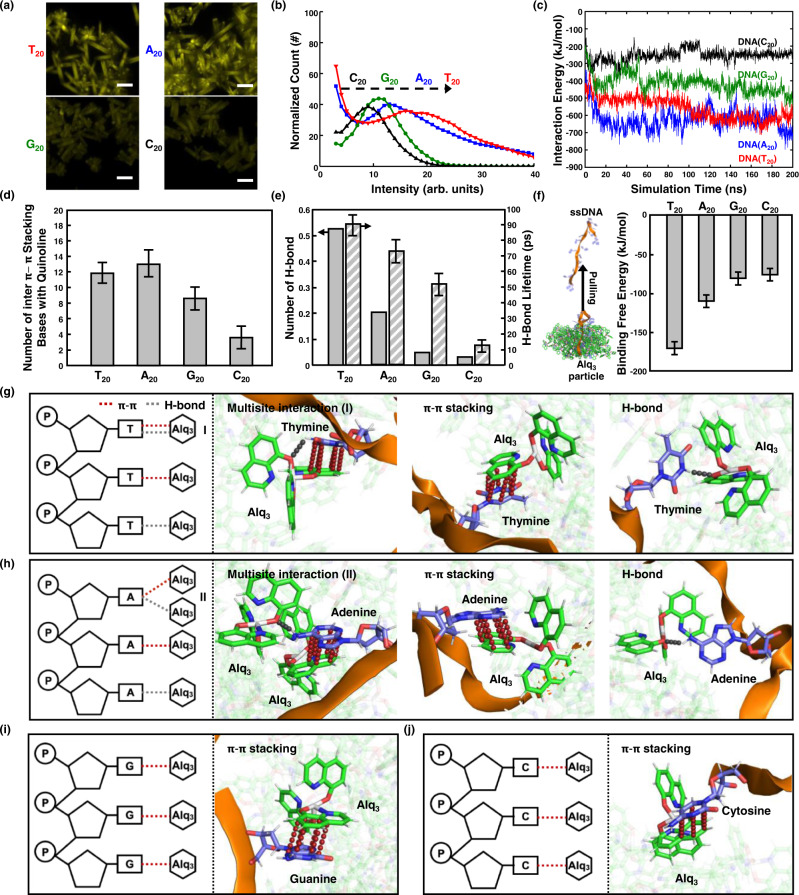
Fig. 3. Effect of base units on molecular doping of DNAs possessing distinct intermolecular interactions.
a Fluorescent analysis of doped DNA(T20, A20, G20, and C20)-Cy3 in Alq3 particles. CLSM images are obtained with excitation by a 555 nm laser (scale bar, 10 μm). b Quantitative histogram of doped DNA-Cy3 using CLSM. Molecular dynamics simulations for the base-dependent incorporation of DNAs into Alq3 microparticles. c Time-traced total interaction energy between Alq3 particles and DNAs. d Number of bases that form inter π-π stacking with the quinoline of Alq3 assembly during simulation. e Number of hydrogen bonds formed between Alq3 molecules and DNAs and their corresponding lifetimes during the final 100 ns simulation. f Binding free energy of the DNAs on Alq3 assembly, obtained from umbrella sampling for three replicates of DNAs fully adsorbed on Alq3 particles. The schematics and molecular views of (g) T20, (h) A20, (i) G20, and (j) C20 doped in Alq3 particles from the final 10 ns of AA MD simulation. Three key interactions, including multisite interaction, stoichiometric π-π stacking, and hydrogen bonding, for doping of T20 and A20 into Alq3 are shown in (g) and (h), respectively. π-π stacking interaction doping G20 and C20 into Alq3 are shown in (i) and (j), respectively. The hydrogen bonds and π-π stacking interaction in schematics and molecular views are depicted in gray and red dashed lines, respectively. Error bars in (d), (e), and (f) represent standard deviation. Source data are provided as a Source Data file.