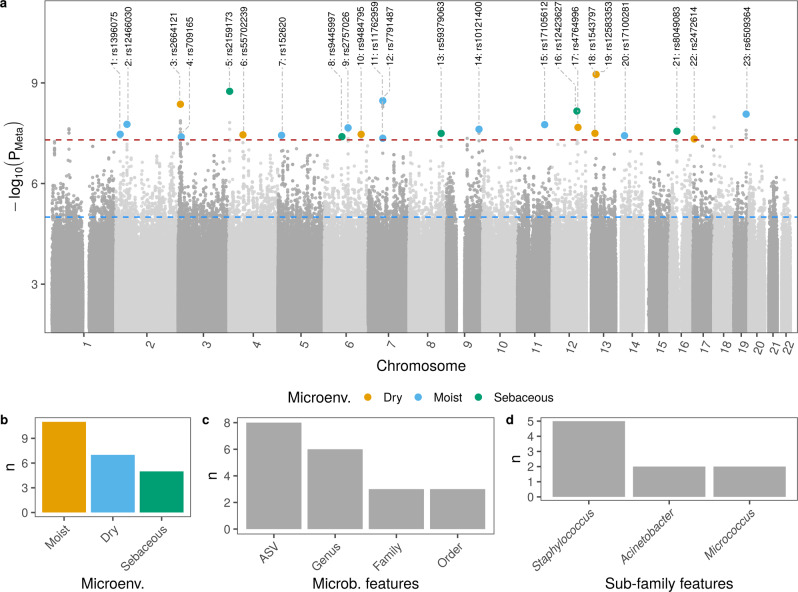
Fig. 2. Results from the GWAS.
a Manhattan plot of per skin microenvironment meta-analysis. Lowest P value of each position is shown and identified by locus ID and rsID. Meta-analysis P values were obtained using the software METAL and METASOFT or by combining P values from data sets that originated from dry skin sites, see Methods. Significant positions are colored according to skin microenvironment and listed, where leading genetic variant, protein coding genes selected by fine-mapping as containing possible causal variants and microbial features are reported. Table 1 contains the list of loci characteristics and genes. b Count of significantly associated loci per microenvironment. c Level of microbial features with highest number of significant associations. d Sub-family features with the highest number of significant associations.