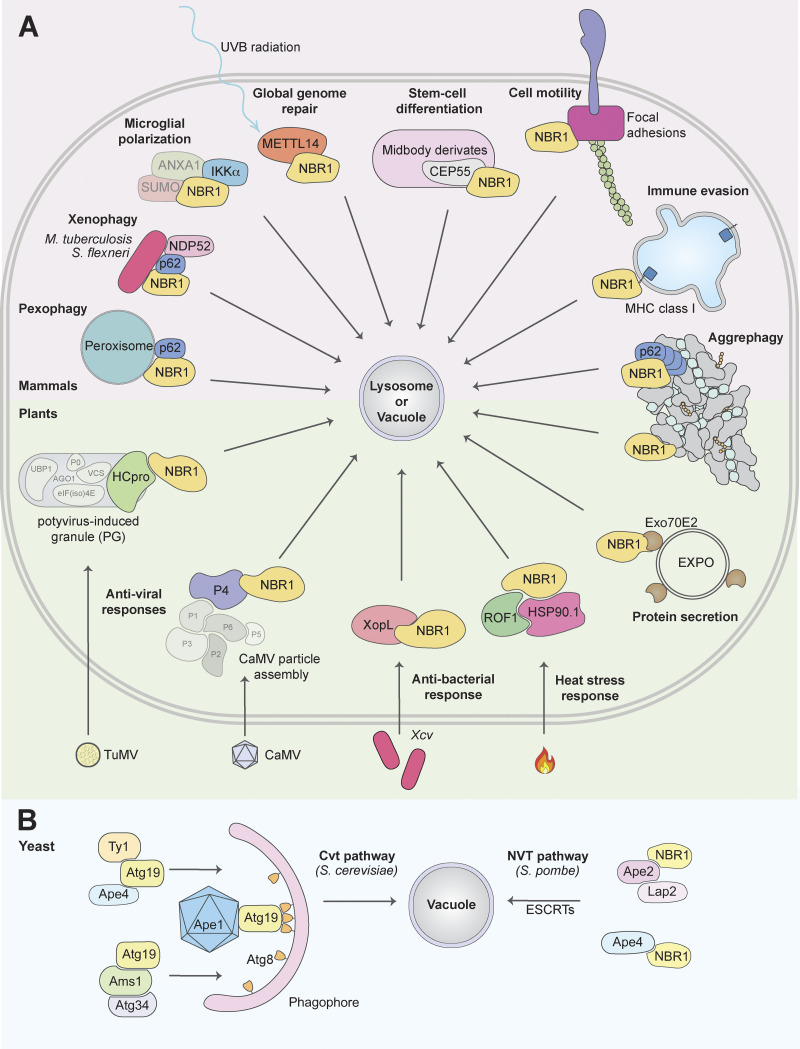
Figure 1.
NBR1 as a selective autophagy receptor in mammals, plants, and yeast. Summary of identified substrates of NBR1-mediated selective autophagy, indicated by immediate vicinity to NBR1. This does not distinguish direct or indirect interaction. Transparent proteins are part of a complex. (A) In mammals, NBR1 has been shown to mediate the degradation of peroxisomes (pexophagy), bacteria (xenophagy), and protein aggregates (aggrephagy) in conjunction with p62 (and NDP52 for xenophagy). Furthermore, NBR1 has been shown to affect several processes through selective autophagic degradation of the following substrates: the proinflammatory kinase IKKα, affects microglial polarization following ischemia (Li et al., 2021); METTL14 (methyltransferase-like 14) upon ultraviolet B radiation, consequently affecting global genome repair (Yang et al., 2021); the midbody protein CEP55 upon stem-cell differentiation (Kuo et al., 2011); turnover of focal adhesions, promoting cell motility; and MHC class I proteins in PDAC cells, promoting immune evasion. In plants, NBR1 regulates several plant stress responses: clearance of aggregates (aggrephagy), restricting TuMV infection by targeting the viral RNA silencing suppressor component HCpro; restricting CaMV infection by targeting the viral particle protein P4; targeting the bacteria effector protein XopL for degradation, restricting Xcv infection; promoting heat stress recovery by targeting ROF1 and HSP90.1; targeting Exo70E2, a marker for the exocyst-positive organelle (EXPO; Ji et al., 2020). (B) In S. cerevisiae, the NBR1 homolog Atg19 mediates the degradation of Ty1, Ape4, Ape1, and Ams1 through the Cvt pathway. The second NBR1 homolog Atg34, only targets Ams1. In S. pombe, the NBR1 homolog targets Ape2, Lap2, and Ape4 to the vacuole. This Nbr1-mediated vacuolar targeting (NVT) pathway is mediated by ESCRTs, not macroautophagy. Only references not cited in the main text are cited here.