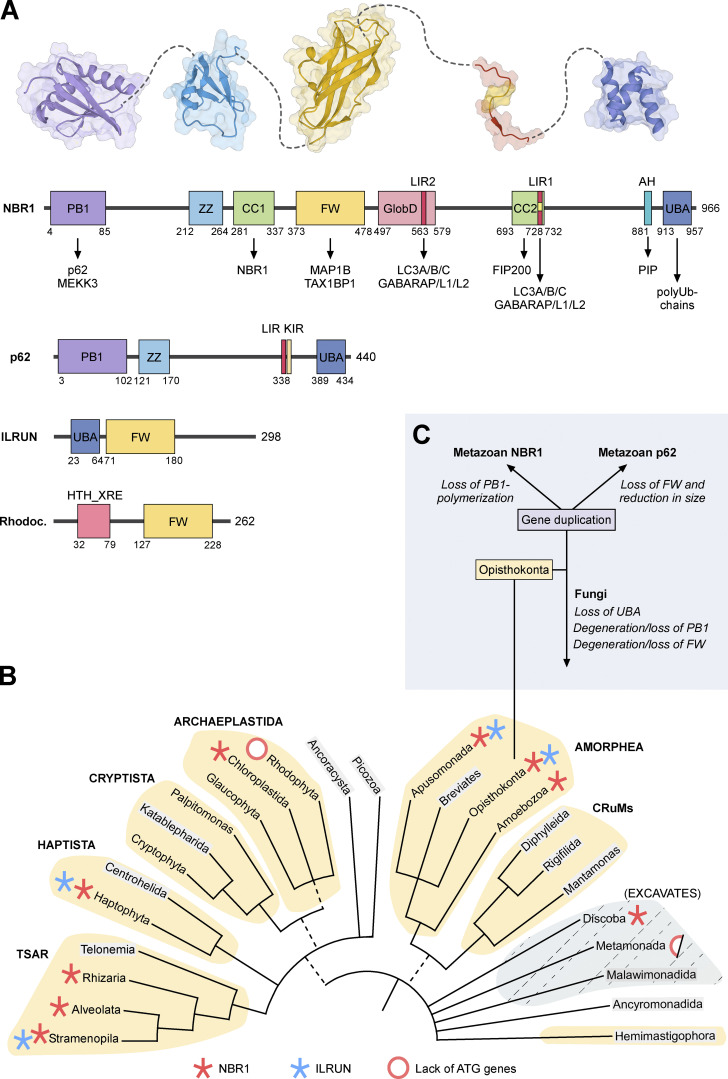
Figure 2.
Domain structure and evolution of NBR1. (A) Domain architectures of human NBR1, p62, and ILRUN, and HTH-XRE (Helix-turn-helix XRE-family like protein) from Rhodococcus fascians. The amino acid positions of the domain borders and the length of the proteins are indicated by numbers below and to the right of the cartoons, respectively. Structures of PB1, ZZ, FW, LIR, and UBA domains are shown above the NBR1 domain architecture. (B) Distribution of NBR1 (red asterisk) and ILRUN (blue asterisk) on The New Tree of Eukaryotes. Red ring, or half-red ring, indicates the lack of ATG genes in some clades. The colored groupings are the current “supergroups.” Multifurcations indicate unresolved branching orders among lineages while broken lines represent minor uncertainties about the monophyly of certain groups (Burki et al., 2020). We searched the NCBI Protein Database and the Conserved Domains Database to identify NBR1 and ILRUN homologs (Lu et al., 2020). The phylogenomic distribution was also determined using the SMART database (Letunic et al., 2006). The names of the groupings where sequence data were not available are indicated in gray. (C) The Ophistokonta contains both the metazoans and fungi. The gene duplication and divergence of the ancestor NBR1 gene in the early metazoan lineage, and loss of UBA and loss or degeneration of PB1 domains in fungi are indicated.