FIGURE 1.
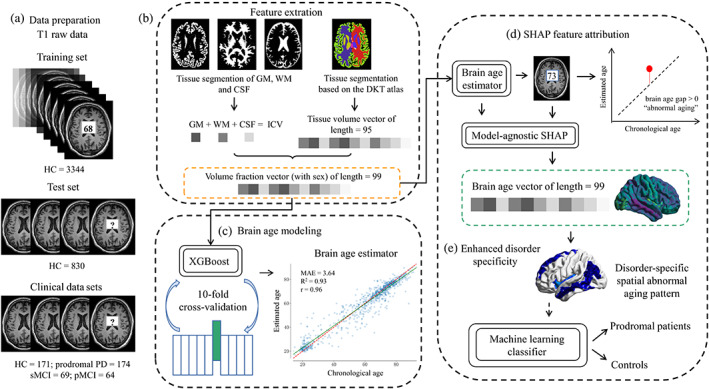
The brain age vector framework, including (a) data preparation. (b) Feature extraction: 95 volumetric features based on DKT atlas were extracted using FastSurfer, and the other three volumetric features of GM, WM, and CSF were extracted by SPM12. All brain volumes were normalized by ICV. (c) Brain age modeling: A total of 98 volume fractions and sex info were used to construct the brain age model based on XGBoost algorithm. Ten‐fold cross‐validation was carried out to tune the model parameters, and the whole training set was used to train the final brain age estimator with the optimal parameters. (d) SHAP feature attribution: We used SHAP to attribute brain aging to each brain region by calculating feature contributions (Shapley values) for each age estimation in the clinical data sets, and the 99 calculated Shapley values formed the brain age vector. (e) Enhanced disorder specificity: The brain age vector reflected disorder‐specific spatial abnormal aging patterns and was further used to classify prodromal patients and controls based on machine learning methods. CSF, cerebrospinal fluid; DKT, Desikan‐Killiany‐Tourville; GM, gray matter; HC, healthy control; ICV, intracranial volume; PD, Parkinson disease; pMCI, progressive mild cognitive impairment; SHAP, Shapley Additive Explanations; sMCI, stable mild cognitive impairment; WM, white matter
