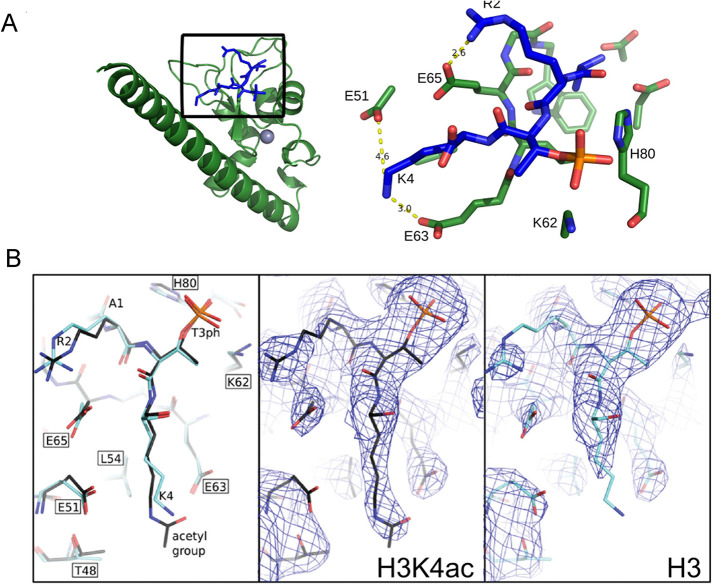
FIGURE 1:
Histone H3K4ac disrupts salt bridges between H3K4 and Survivin E51 and E63. (A) Structure of human Survivin (green) bound to the unacetylated histone H3 peptide (blue). Gray sphere represents zinc atom. Inset shows amino acids involved in interaction between the H3 peptide and the Survivin to highlight the binding interface between Survivin and the histone H3T3phK4 peptide; yellow dashed lines indicate formation of salt bridges between histone H3 lysine 4 side chain and Survivin glutamic acids E51 and E63. Survivin carbon, nitrogen, and oxygen atoms are shown as green, blue, and light red sticks, respectively. Histone atoms are shown in blue; phosphorous and oxygen atoms of phosphorylated T3 are colored orange and red, respectively. (B) Comparison of Survivin bound histone H3 peptide with and without K4 acetylation. Left, stick representation showing unacetylated (light blue) and acetylated (dark blue) peptides; center and right, electron density in the vicinity of Survivin histone H3 peptide binding site with the view on unmodified and acetylated lysine 4. The density around δ and ε carbon of lysine 4 is absent because unmodified K4 side chain can adopt many conformations including salt bridge formation between E51 or E63; therefore it appears disordered in the model. After K4 acetylation there are less possible conformations that can be adopted by acetylated K4 side chain due to steric effects; therefore lysine side chain appears ordered in the model. Map is contoured at 1 sigma.