Fig. 1.
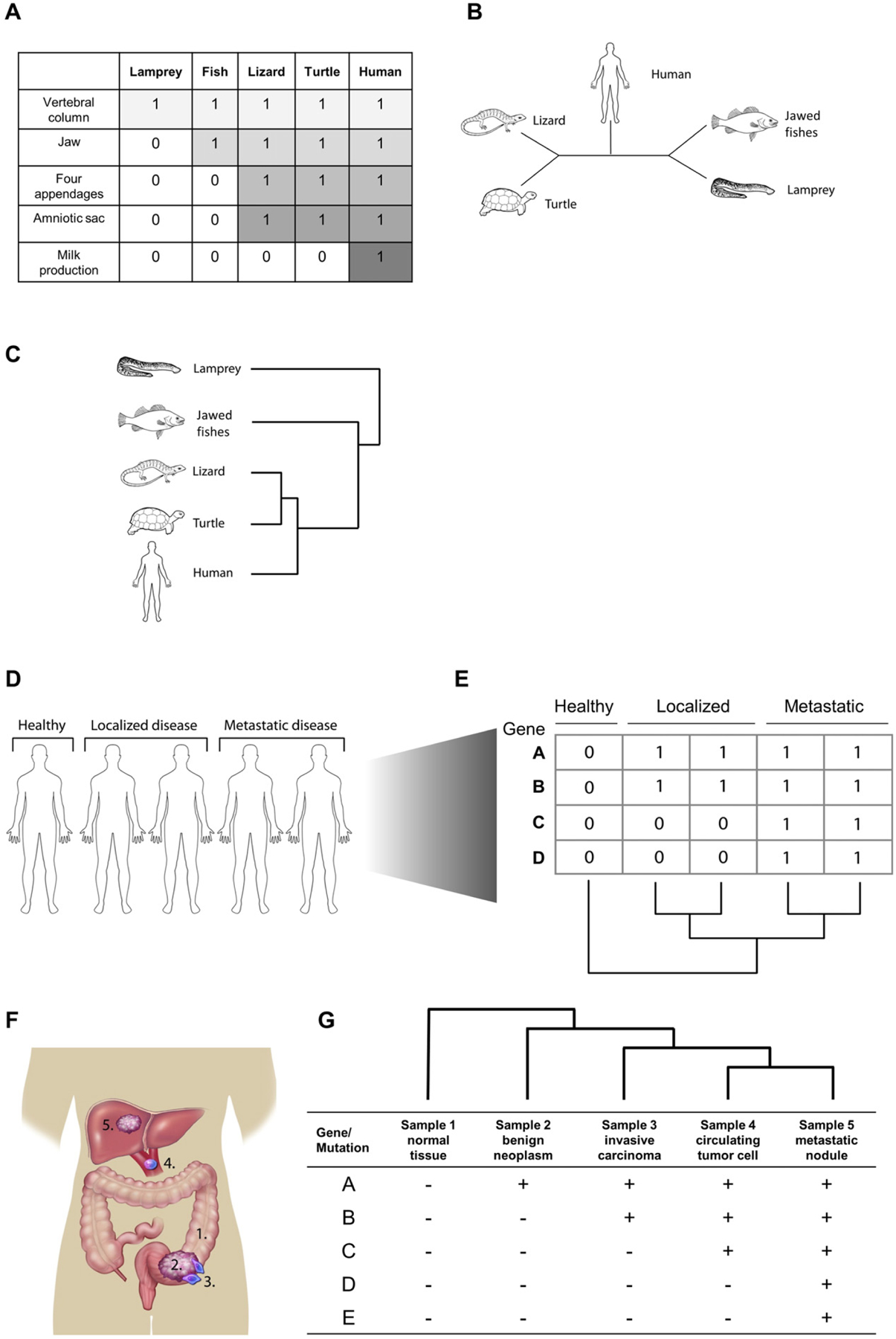
Phylogenetics reveals evolutionary relationships between states. A. Characteristics from various species under study can be transformed into a binary character state matrix. A species that is known to possess the ancestral state of the given characters (e.g. here, the lamprey) can be included as an “outgroup” as a means by which to polarize the resulting tree. B. An unrooted most parsimonious tree obtained by choosing the topology requiring the fewest number of character changes. C. The unrooted tree is converted to a rooted tree by assuming that jawed vertebrates share a more recent common ancestor than the most recent common ancestor (MRCA) of the entire group. C. As a cancer research tool, phylogenetic analyses can be used strictly as a clustering algorithm to segregate individual patients by their progression status. D. Samples are collected from individual patients, a matrix of characters is constructed using gene expression, mutation status, or some other information, and a phylogenetic tree is generated. E. In a more direct application of phylogenetic methods, they can be used to analyze phenotypic/genotypic heterogeneity within a patient or disease location. In this example, samples are collected at different sites to construct a matrix and tree of progression F. Depending on the question being asked, samples can be collected longitudinally or from neighboring areas of a tissue a single site (e.g. primary tumor and metastatic nodules) to reconstruct the evolutionary history of the disease progression.
